Salmonella Typhimurium (S. Typhimurium) is the most common cause of foodborne illness worldwide but can be difficult to characterize. Researchers at Tennessee State and Auburn Universities have addressed this issue in their 2019 article published in the journal Antibodies. Both Surface Plasmon Resonance (SPR) and enzyme-linked immunosorbent assays (ELISA) have been used for this type of analysis, but the sensitivity and reliability of the assays have been problematic. These researchers are working to improve this situation by characterizing monoclonal antibodies (Mabs) used in these assays more thoroughly. They employed a Reichert SR7500DC to characterize S. Typhimurium interactions with various antibodies.1
Looking more closely at the S. Typhimurium bacterium, one finds it has 6–10 flagella with 3 substructures. One of the substructures, the filament, extends into the extracellular space and is comprised of a single species of protein called flagellin. Further, S. Typhimurium has two non-allelic genes that encode two antigenically distinct flagellins, and monoclonal antibodies can bind to one or the other depending on the specificity of the antibody. The development of numerous immunoassays (ELISA and SPR) based on flagellin, including the current SPR research outlined here, have been made straightforward because: (a) a number of different anti-flagellin antibodies are available, and (b) that Salmonella flagellin can be extracted fairly easily.1
Experimental
Background
An anti-flagellin antibody was immobilized on a dextran chip using standard amine coupling. Unfilled sites were blocked with BSA. Flagellin was injected over the surface and captured and then binding to either one Mab, or two Mabs in a row, was monitored to determine kinetics and carry out epitope mapping, respectively.
A sandwich type assay has been employed to study S. Typhimurium. The capture antibody used is Mab 1E10. This was chosen because MAb 1E10 was found to have a different epitope compared to the rest of the antibodies that were tested. For kinetics alone, flagellin was captured and then four concentrations of each antibody were injected over the surface and the curves were globally fit to obtain rate constants. Next, epitopes were determined. After flagellin capture, an initial antibody followed by a second antibody were injected to determine whether or not inhibition occurred. In addition, responses were quantified by looking at Bmax ratios.1
Conditions
- Instrument: SR7500DC/Reichert2SPR
- Sensor Chip: Dextran
- Running Buffer: PBST
- Flow Rate: 20 μL/min
- Temperature: 25°C
- Capture Antibody: MAb 1E10 (amine coupled at pH 5.2)
- Analyte: Salmonella flagellin (4 minute association, 6 minute dissociation)
- For kinetics: Antibody 1 is injected over the analyte (4 minute association, 6 minute dissociation)
- For epitope mapping: After flagellin, 2 Mabs in a row are injected (Antibody 1 and Antibody 2) (4 minute association, 4 minute dissociation each).
- Regeneration Buffer: 10 mM Glycine-HCl, pH 3.0 (4 min injection)
Results
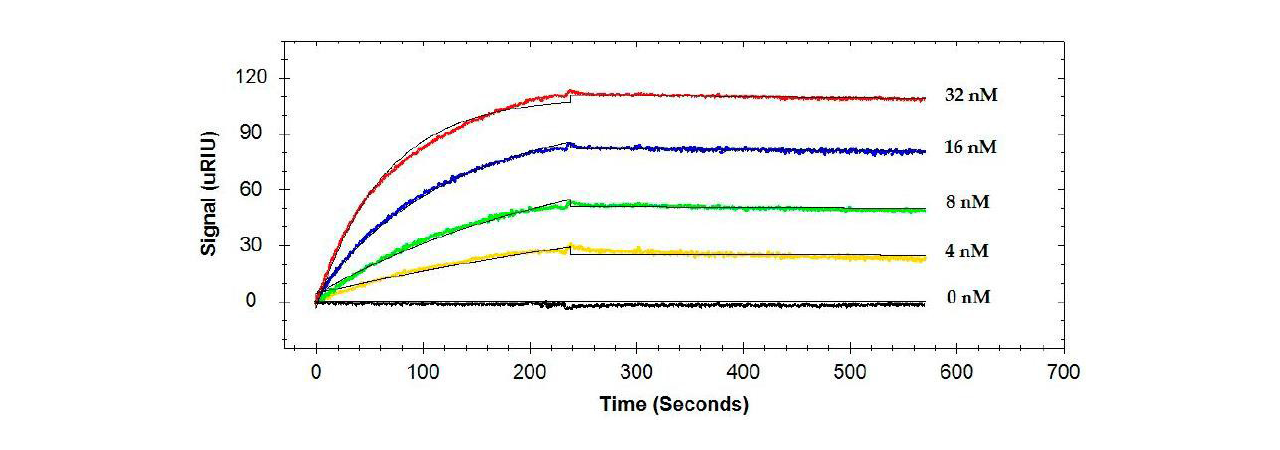
Binding of single antibodies to the flagellin was, in general, high affinity with KD values ranging from 26.3 to 325 pM (see example in Figure 1).1
For epitope testing, when different combinations of two antibodies were injected consecutively, some antibodies had no effect on each other (bound to different sites as in Figure 2 (a)) and others inhibited (had a shared binding site as in Figure 2 (b)) or enhanced the response of the secondary antibody.1
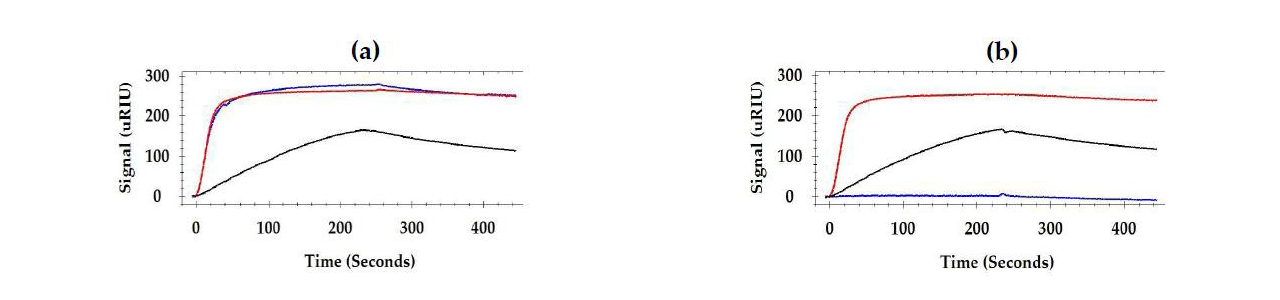
Summary
- A Reichert SPR was used to characterize the binding of monoclonal antibodies to flagellin from the Salmonella Typhimurium bacterium. Due to the ability to capture the flagellin in an oriented manner, SPR was found to offer greater sensitivity compared to ELISA.1
- With the epitope-captured-flagellin approach, four distinct epitopes were identified for the five monoclonal antibodies. The four epitopes are located within a 23 kDa domain of flagellin (50 kDa).1
- There are several cases where enhancement of the binding of the secondary antibody was observed. This may be attributed to the differences in the affinities between the two antibodies and the conformational changes of flagellin caused by the binding of the first antibody. Other epitope testing resulted in less or even no binding of the secondary antibody.1
Reference
- Devendra Bhandari, Fur-Chi Chen, Shreya Hamal and Roger C. Bridgman, “Kinetic Analysis and Epitope Mapping of Monoclonal Antibodies to Salmonella Typhimurium Flagellin Using a Surface Plasmon Resonance Biosensor”, Antibodies, 2019, 8(1), 22; doi:10.3390/antib8010022