Introduction
Researchers at the University of Pittsburgh have been studying an HIV-encoded protein called Nef, a so-called accessory protein that facilitates the process of HIV/AIDS development. Prior research has shown that Nef plays a critical role in the viral life cycle and the host immune response to the infection, and that certain protein-protein interactions are critical to these Nef functions. Interactions with the SH3 or tandem SH3-SH2 domains of the Src-family of kinases are examples of the critical protein-protein interactions. X-ray crystallography has revealed that distinct dimer conformations of Nef result from interaction with the SH3 or tandem SH3–SH2 domains of the Src-family kinase known as Hck. By studying these interactions in solution, University of Pittsburgh researchers have been able to characterize the kinetics and affinities of these interactions, and they have been doing so using Reichert Surface Plasmon Resonance (SPR) instruments. SPR has primarily been used to provide corroboration of hydrogen exchange mass spectrometry (HX MS) results which support a model in which Src-family kinase binding induces conformational changes in Nef in solution, which then exposes residues critical for subsequent interactions related to MHC-I downregulation and immune escape of HIV-infected cells.1
Treatment with antiretroviral drugs has become the accepted approach to HIV/AIDS therapy. Drugs are usually taken in combinations that include nucleoside reverse transcriptase inhibitors, which block how the HIV genetic material is used to create DNA from RNA, protease inhibitors, where the raw material for new HIV virus is cut into specific pieces, and integrase inhibitors, which prevent the proviral HIV DNA from integrating into the host cell genome. These drugs do not provide a cure but can help slow the disease's progression.2 Researchers in this current study ultimately would like to develop a new class of antiretrovirals that can interfere specifically with the functions of HIV1/Nef. Since Nef is critical for HIV-1 replication in vivo and the immune escape of HIV-infected cells which leads to AIDS progression, development of potent HIV-1/Nef inhibitors hold promise in the suppression of HIV replication and the restoration of immune recognition of HIV-positive cells so that the patient's own CTL response can be used to combat the disease.1 In this current work, HX MS and SPR have been the primary techniques used to better understand the mechanism of action of these kinases on Nef conformational transitions in solution. Hence, this study supports the broader possibility that compounds interfering with conformational transitions in HIV-1/Nef structure have potential as a new class of antiretroviral agents.1 This is important as viruses can become resistant to existing treatments over time and new treatments will continue to be needed
Experimental
Background
Nef is a small protein with a molecular weight of 27-35 kDa which interferes with the cell-surface display of major histocompatibility complex-1 (MHC-1)/HIV antigen complexes on infected cells, thereby evading HIV-specific cytotoxic T cells.1,3 Nef binds and activates several non-receptor protein-tyrosine kinases, including members of the Src family. Inhibition of these Nef-activated kinases has been shown to suppress Nef-dependent enhancement of HIV-1 infectivity and replication, suggesting that these kinases activate downstream signals that promote the viral life cycle. Src-family members expressed in HIV-1 target cells, including Hck, are also required for Nef-dependent MHC-1 downregulation.1
To better understand the interaction between Hck kinases and Nef in solution, recombinant Hck SH3 and SH3–SH2 proteins were amine coupled to dextran chips. Nef proteins were then flowed over the chip surface at concentrations ranging from 0.041–3.3 μM. Three types of Nef proteins were studied: full-length wild-type Nef, a truncated Nef core protein lacking the N- terminal anchor region, and a dimerization defective Nef core mutant, Nef-D123N. Kinetics were determined using Tracedrawer.1
Conditions
- Instrument: SR7500DC/Reichert2SPR
- Sensor Chip: Dextran
- Running Buffer: 10 mM Hepes (pH 7.4), 150 mM NaCl, 3 mM EDTA, 0.05% v/v Surfactant P20 (+ 1 mM DTT)
- Flow Rate: 30 μL/min
- Association Time: 1 minute (until equilibrium reached)
- Dissociation Time: 3 minutes
Results
Researchers were interested in knowing whether binding properties changed depending on whether the Nef protein was full-length or only the folded core lacking the N-terminal arm responsible for membrane anchoring (amino acids 1-60) was present. They also investigated a mutant defective for homodimer formation, in which Nef Asp123 was changed to asparagine (Nef-D123N). Substitution of this highly conserved aspartate prevented dimerization in cell-based fluorescence complementation assays and blocked important Nef functions related to infectivity and replication, as well as CD4 and MHC-I downregulation.1
After immobilization of SH3 or SH3-SH2 ligand, Nef protein was flowed over the chip either as full- length analyte, wild type core or D123N core. Examples of responses seen for the wild type Nef core are shown below in Figures 1 and 2. When SH3 alone was coupled, the wild type Nef core exhibited single-site binding with a KD value of of 128 nM. Binding was similar for the other Nef proteins - the affinity was a little lower for the full-length Nef (180 nM) and a little higher for the D123N core (119 nM). When SH3-SH2 was employed as the ligand, 2 site binding (conformational change) was seen with an affinity of 475 nM for the Nef core and similar affinities for the other Nef proteins (429 nM and 475 nM for the full-length Nef and D123N core, respectively). Hence, it was found that the mutation had little impact on the kinetics or affinity of SH3 domain interaction, suggesting that the D123N mutation does not influence the conformation of Nef to inhibit binding. This finding was consistent with HX MS results.1
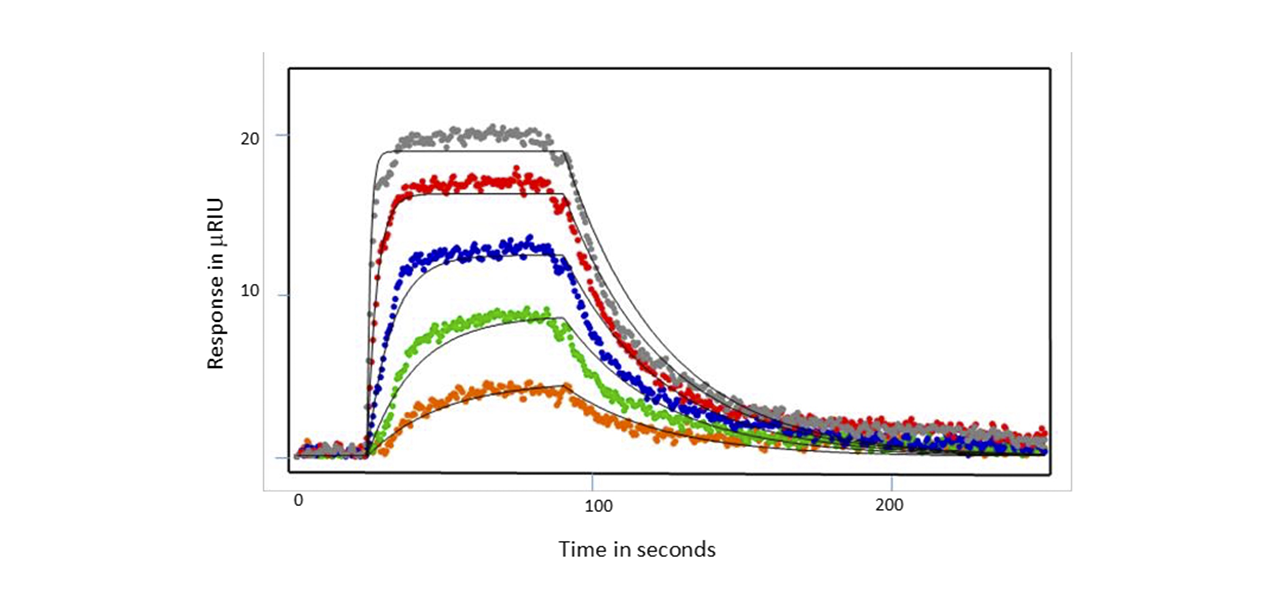
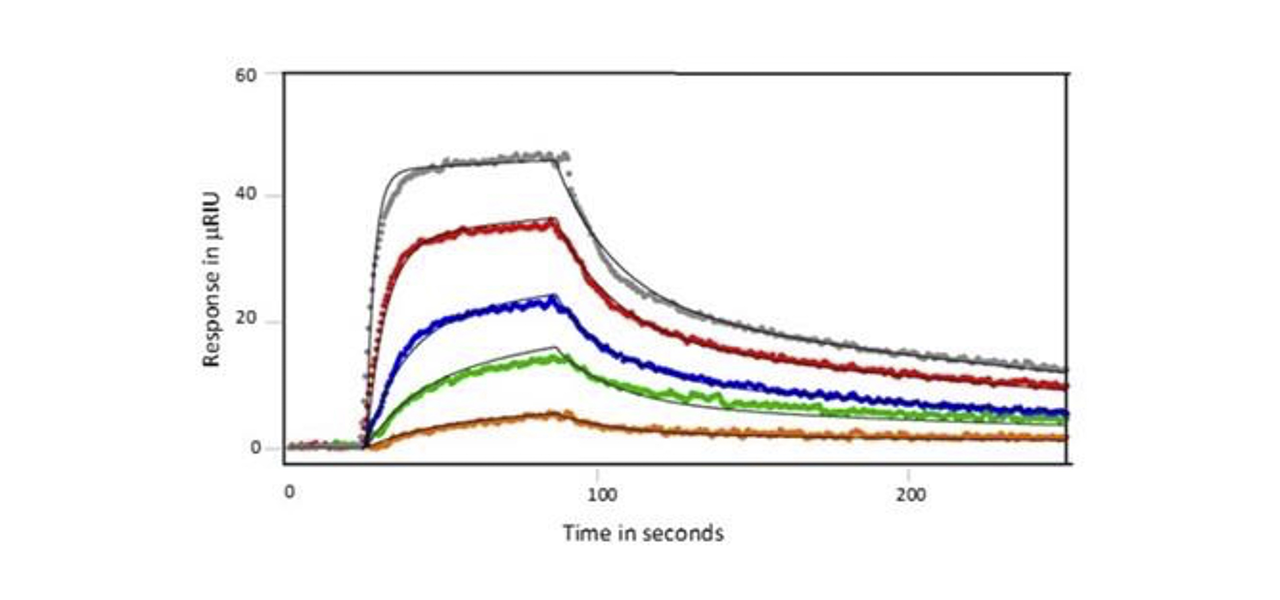
Summary
- SPR provided quantitative kinetics for the interactions between Nef and two regulatory domain proteins (SH3 and SH3-SH2) derived from a kinase linking Nef and HIV replication. SPR rate constants were independent of whether the full length Nef or core molecules were tested.
- A 1:1 binding model fit best with SH3 as the target compared to a conformational change model when SH3-SH2 was the target, consistent with previous structural work showing that the SH3-SH2 protein forms extended contacts with Nef, resulting in more compact structure compared to the SH3-only complex.
- SPR results were consistent with findings from hydrogen exchange MS, which showed that SH3 and SH3-SH2 interaction with Nef occurs at a site distinct from the Nef dimerization interface.
- These findings will help drive further research to develop specific molecules to target Nef.
References
- Jamie A. Moroco, John Jeff Alvarado, Ryan P. Staudt, Haibin Shi, Thomas E. Wales, Thomas E. Smithgall, John R. Engen, "Remodeling of HIV-1 Nef Structure by Src- Family Kinase Binding," Journal of Molecular Biology, Available online 16 December 2017, ISSN 0022-2836, doi.org/10.1016/j.jmb.2017.12.008.
- Aidsinfonet.org Fact Sheet 403: What Is Antiretroviral Therapy (ART)?
- Pawlak, E. N.; Dikeakos, J. D., “HIV-1 Nef: a master manipulator of the membrane trafficking machinery mediating immune evasion,” Biochim. Biophys. Acta, 2015, 1850 (4), 733-741.