Introduction
The accumulation of extracellular plaques containing the neurotoxic ß-amyloid peptide fragment, Aß(1-42) of ßamyloid precursor protein (ßAPP), is one of the characteristics of Alzheimer's disease (AD). Although ßAPP has been recognised as a key molecule for AD, its molecular (patho) physiological degradation, proteolytic pathways and cellular interactions of Aß are still unclear. Studies towards the development of immunotherapeutic methods for AD have yielded initial success in transgenic mouse models of AD and in producing therapeutic antibodies by immunization with Aß(1-42) that disaggregate Aß-plaques and fibrils. Using proteolytic excision of the immobilized Aß antigen-immune complex in combination with ESI mass spectrometry, the Aß-plaque specific epitope was identified as an N-terminal peptide Aß(4-10), accessible in Aß(1-42) as well as in oligomeric Aß-fibrils [1, 2].
Recently, we have identified the epitope recognized by Aß-autoantibodies in serum, capable of eliciting a neuroprotective effect to inhibit the formation of Aß-plaques, located in the carboxy terminal region of the Aß sequence. The differential epitope structures of Aß-specific antibodies from healthy individuals and AD patients provides a breakthrough and molecular basis for (i), the development of new immuno-therapeutic approaches by passive immunization with Aß-specific antibodies, and (ii), the development of new diagnostic tools for AD with absolute specificity [2, 3]. The primary structures of polyclonal Aß-autoantibodies were elucidated by two-fold Aßepitope specific affinity chromatography from human immunoglobulin G, using a combination of overlapping proteolytic digestion (trypsin, α-chymotrypsin), HPLC isolation, and high resolution MS, which provided sequence data for Fv domains, CDR motifs, and framework regions.
The new online SPR-MS system is capable of detecting and identifying affinity interactions in real time. The reliability of the online interface was established by demonstrating repeatable and comparable KD determinations and precise mass spectrometric identification of protein. Fast on-line sample processing allows fast throughput of different analytes for biomolecular interaction studies. In this study, the plaque protective Aß-epitope of the Aß-autoantibody was identified by proteolytic extraction-MS using the online SPR-MS- epitope analyser, and found to reside in the Aß(17-28) tryptic peptide sequence.
Experimental
To identify the epitope, the Aß- autoantibody was immobilized on a dextran-SPR affinity chip, and a tryptic mixture of Aß-peptide fragments was injected through the SPR autosampler followed by ESI-MS. Subsequent online desalting of analyte prior to MS was performed after elution of affinity captured Aß-peptide which provided identification of the Aß(17-28) epitope peptide (protonated molecular mass 1324,8). Following elution of undigested Aß(1-40) through the microfluidic interface, SPR affinity determination revealed high affinity with a KD of ca. 3.5 nM (Figures 2 and 3). Based on this epitope peptide, interactions of the plaque-protective Aß-epitope with two fibril inhibiting peptides, cystatin-C and humanin, were evaluated at the molecular level to gain insight into the mode of action of Aßautoantibodies [4].A schematic of the setup is shown in Figure 1.
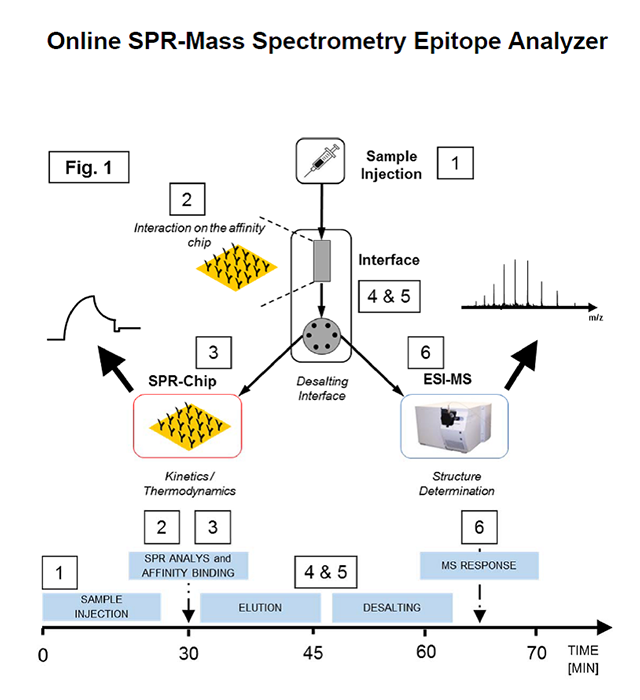
Schematic workflow of epitope & interaction analysis using the online SPR-MS Epitope Analyzer. After sample injection (1), the analyte is captured on the affinity chip (2) followed by the SPR-chip (3) for kinetic analysis of the affinity interaction. After sample processing through the desalting interface (4 and (5), structural analysis is performed by ESI-MS. Time scale is represented on the time axis below.
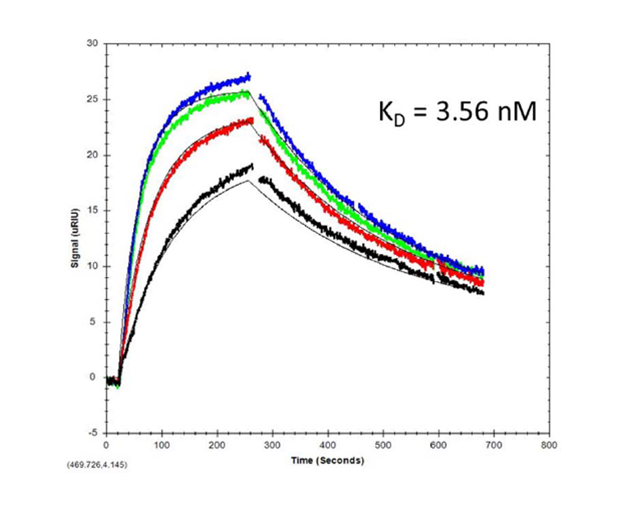
SPR determination of a dilutions series of Aß(1 - 40) upon processing via the SPR-MS interface. Kinetic evaluation resulted in a calculated KD of 3.56 nM.
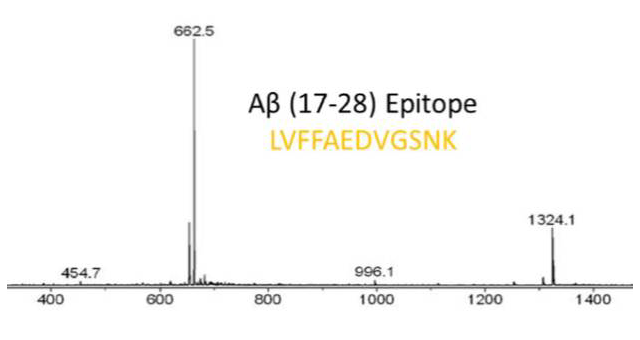
ESI-MS identification by online SPR-MS of the epitope Aß (17-28) eluted from the Aß-antibody upon proteolytic extraction.
Conclusions
In this study we show that the online SPR- ESIMS combination is a powerful tool to enable the simultaneous affinity isolation, structure identification and affinity quantification of an Aß- plaque protective epitope from the complex of Aß-autoantibodies immobilized on a gold chip. The high application potential of online-SPR-MS has become further evident in recent studies of the identification of an unusual mixed-disulfide antibody epitope of the rheumatoic target protein, HLA-B27; and the interaction site identification of chaperone complexes of lysosomal enzymes [5, 6]. Current applications confirm that interaction epitopes as diverse as antigen-antibody and lectin- carbohydrate complexes [7], and binding constants (KD) from milli- to nanomolar ranges are amenable to SPR-MS analysis. These results indicate that applications of the online-SPR-MS epitope analyzer are well feasible to affinity-based biomarker evaluation; identification of protein and peptide epitopes; precise antibody affinity characterization; and direct label-free antigen quantification.
References
- McLaurin, J., R. Cecal, M.E. Kierstead, X. Tian, A.L. Phinney, M. Manea, J.E. French, M.H.L. Lambermon, A.A. Darabie, M.E. Brown, C. Janus, M.A. Chishti, P. Horne, D. Westaway, P.E. Fraser, H.T.J. Mount, M. Przybylski, P. St.-George-Hyslop (2002) Nature Med. 8: 1263-1269.
- Stefanescu R., Iacob R.E., Damoc E.N., Marquardt A., Amstalden E., Manea M., Perdivara I., Maftei M., Paraschiv G., Przybylski M. (2007) Eur. J. Mass Spectrom. 13: 69-75.
- Przybylski, M. et al./Univ. Konstanz & Bonn (2009) Eur. & US Patent Applications; M. Przybylski et al. (2011/2012), Eur. Patent Appl.
- Juszczyk P, Paraschiv G, Szymanska A, Kolodziejczyk AS, Rodziewicz-Motowidlo S, Grzonka Z, Przybylski M. (2009) J. Med. Chem. 52: 2420-2428.
- Iurascu, M.I., Marroquin Belaunzanar, O., Cozma, C., Petrausch, U., Renner C., Przybylski M. (2016) J. Am. Soc. Mass Spectrom. 27: 1105-1112.
- Moise, A., Maeser, S., Rawer, S., Eggers, F., Murphy, M., Bornheim, J., Przybylski, M. (2016) J. Am. Soc. Mass Spectrom. DOI: 1007/13361-016-1386-0.
- Moise A., Andre, S., Eggers, F., Krzeminski, M., Przybylski, M., Gabius, H.J. (2011) J. Am. Chem. Soc. 133, 14844-14847.