Immobilization made easy with Click Chemistry
Probably anyone who has ever performed an SPR experiment was initially faced with the problem of efficient and reproducible ligand immobilization.
The starting point is always the question of which immobilization chemistry is most compatible with the ligand, how to achieve optimal and reproducible ligand densities for a given application, and how to preserve the ligand’s activity. In the end, it’s all about which immobilization strategy generates the highest data quality in your specific SPR experiment.
In this newsletter, we introduce Click Chemistry as a versatile and easy-to-use coupling strategy and point out why Click Coupling is more than just another covalent immobilization method.
Drawbacks of the state of the art
Covalent immobilization via EDC/NHS - the traditional workhorse of ligand immobilization on biosensor chips - has several negative attributes: First, the necessity for a buffer compatible with electrostatic pre-concentration, which is non-physiological, usually with low pH and low ionic strength. Second, the difficulty in controlling the immobilization level, which is hard to forecast in the first experiment and impossible to adjust later on. In addition, there are less obvious factors, such as random coupling sites and cross-linking of the ligand, which may adversely affect the ligand’s binding characteristics.
A complete list of advantages and disadvantages you can find here.
| Covalent EDC/NHS | Covalent click-chemistry | Streptavidin/ biotin | Ni-NTA/ His6 | |
|---|---|---|---|---|
| Non-specific binding | Low | Very low | Moderate | High |
| Baseline drift after immobilization | No | No | Low | Moderate – High** |
| Targeting a specific immobilization level | Difficult | Easy | Easy | Moderate |
| Repeated immobilization cycles | No | Yes | Yes | Yes |
| Pre-concentration mandatory for immobilization | Yes | No | No | No |
| No crosslinking during immobilization | No | Yes* | Yes* | Yes |
| Quenching of reactive groups after immobilization required | Yes | No | No | No |
| Buffer restrictions during immobilization and measurement | Yes | No | No | Yes |
| Excluded volume by functional moiety | Low | Low | Very high |
Low |
*If the conjugation ratio of the protein with the respective label is < 1.
**NiHC moderate, NiD / NTA high.
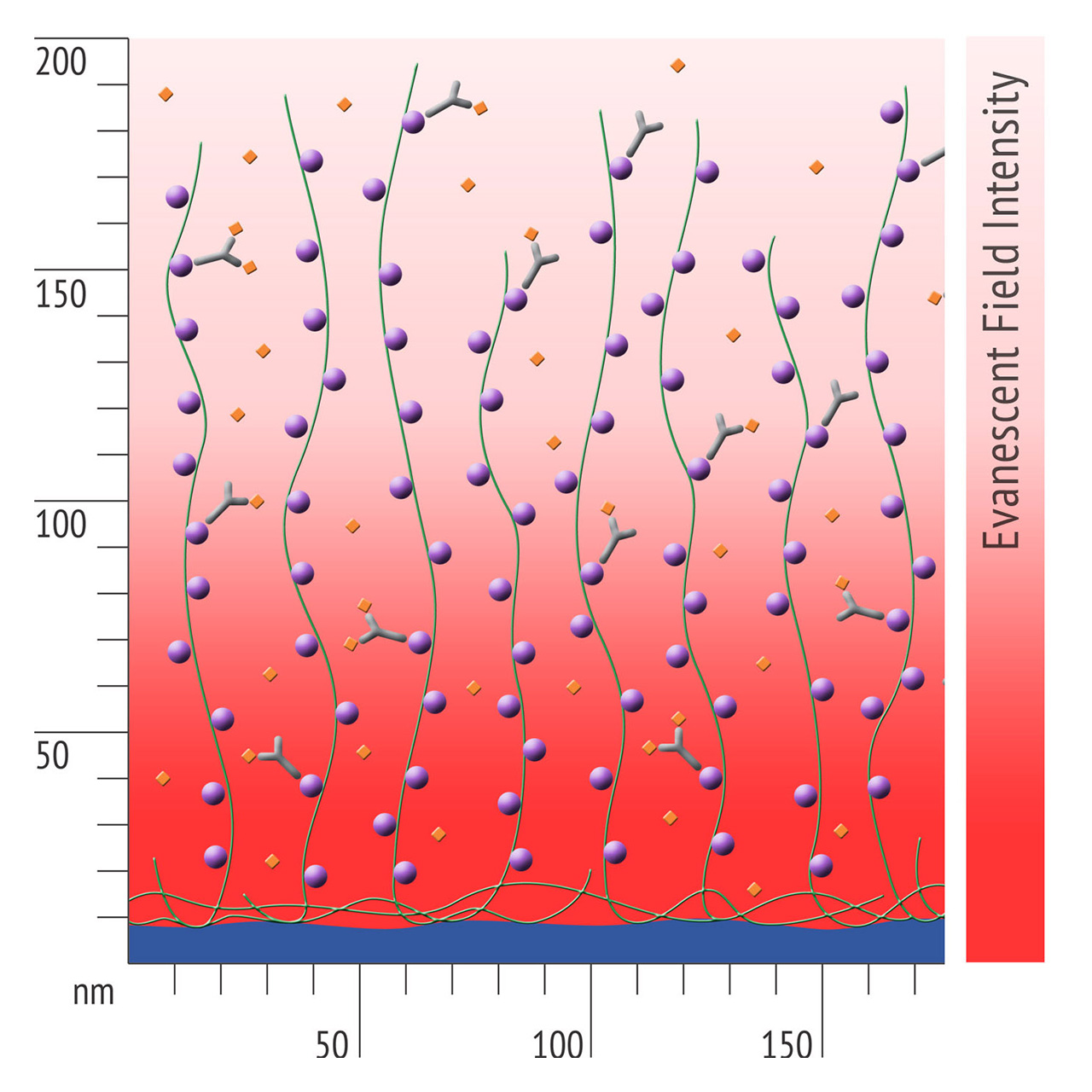
Besides EDC/NHS coupling, affinity based ligand immobilization via streptavidin–biotin linkage has become one of the most popular immobilization methods for SPR biosensing: Though this approach provides some compelling features including convenient handling and excellent immobilization control in physiological conditions, the negative effects of a bulky linker like streptavidin should never be underestimated. Pre-immobilized streptavidin occupies a significant portion of the available space within the evanescent field, resulting in a reduced ligand immobilization yield and entailing the risk of hindered analyte diffusion.
The challenge
Is there an immobilization chemistry which combines the simplicity of biotin–streptavidin immobilization, the immobilization yields of EDC/NHS chemistry, and is in addition highly selective & reproducible, sterically undemanding, and easy to apply?
The solution
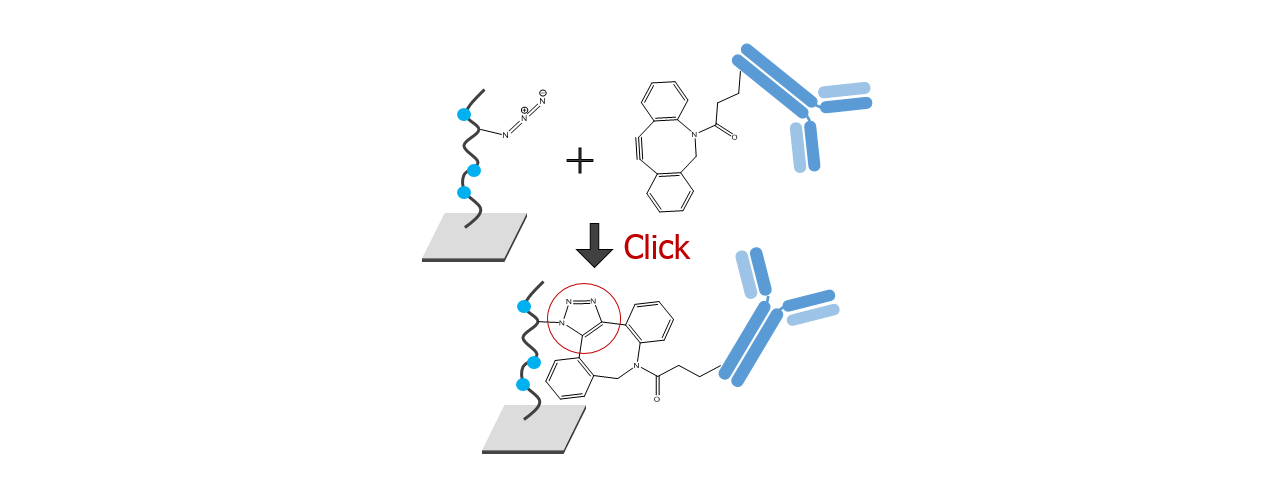
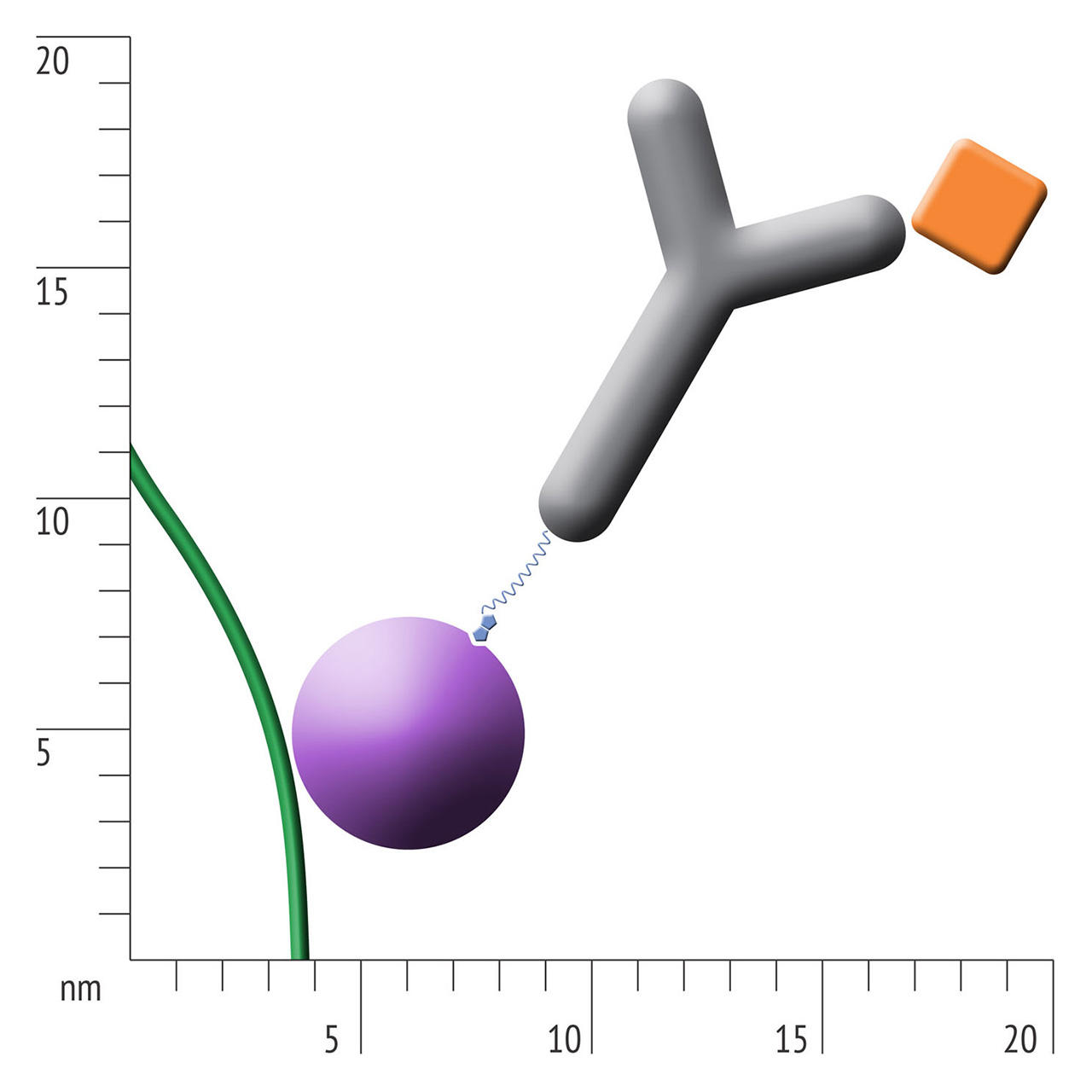
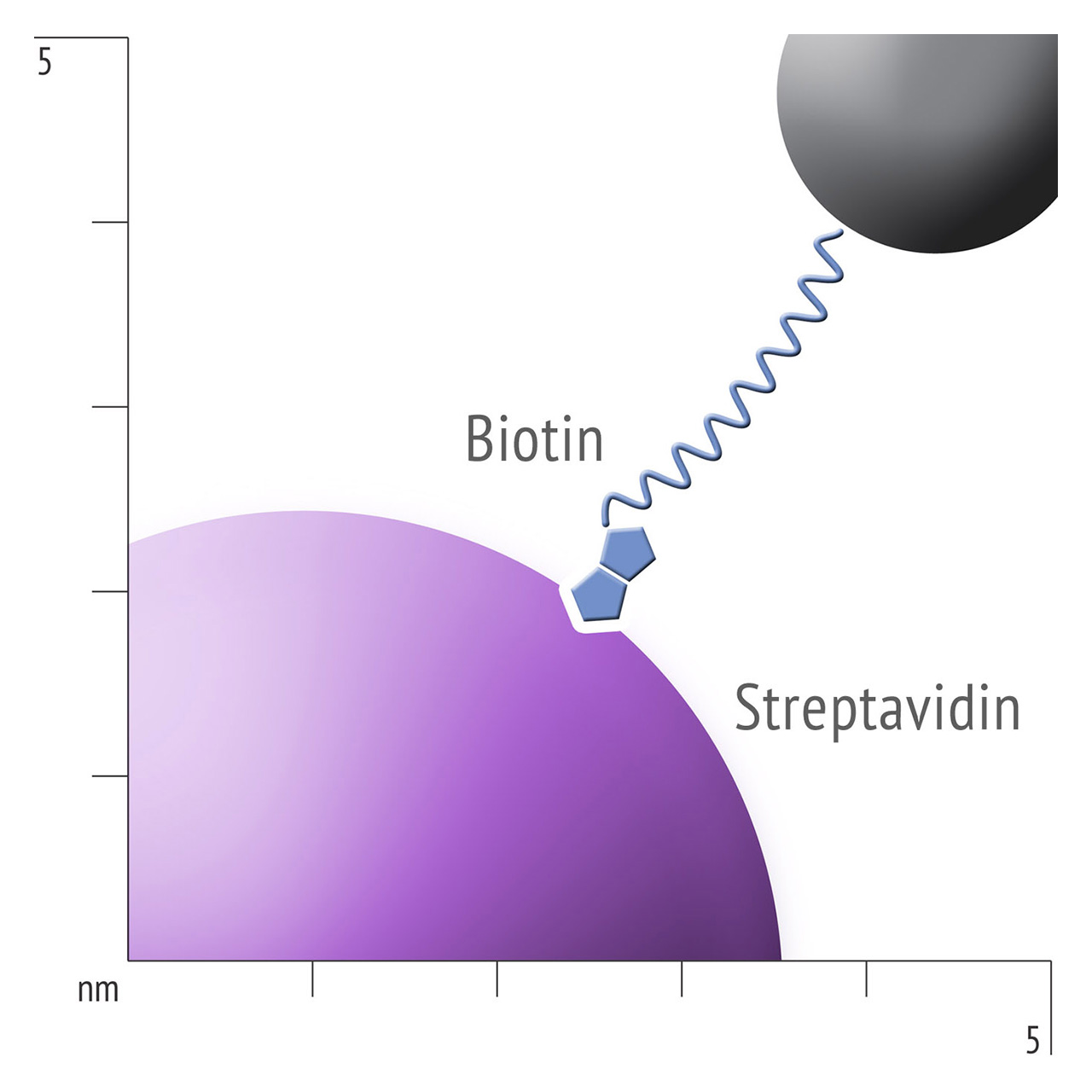
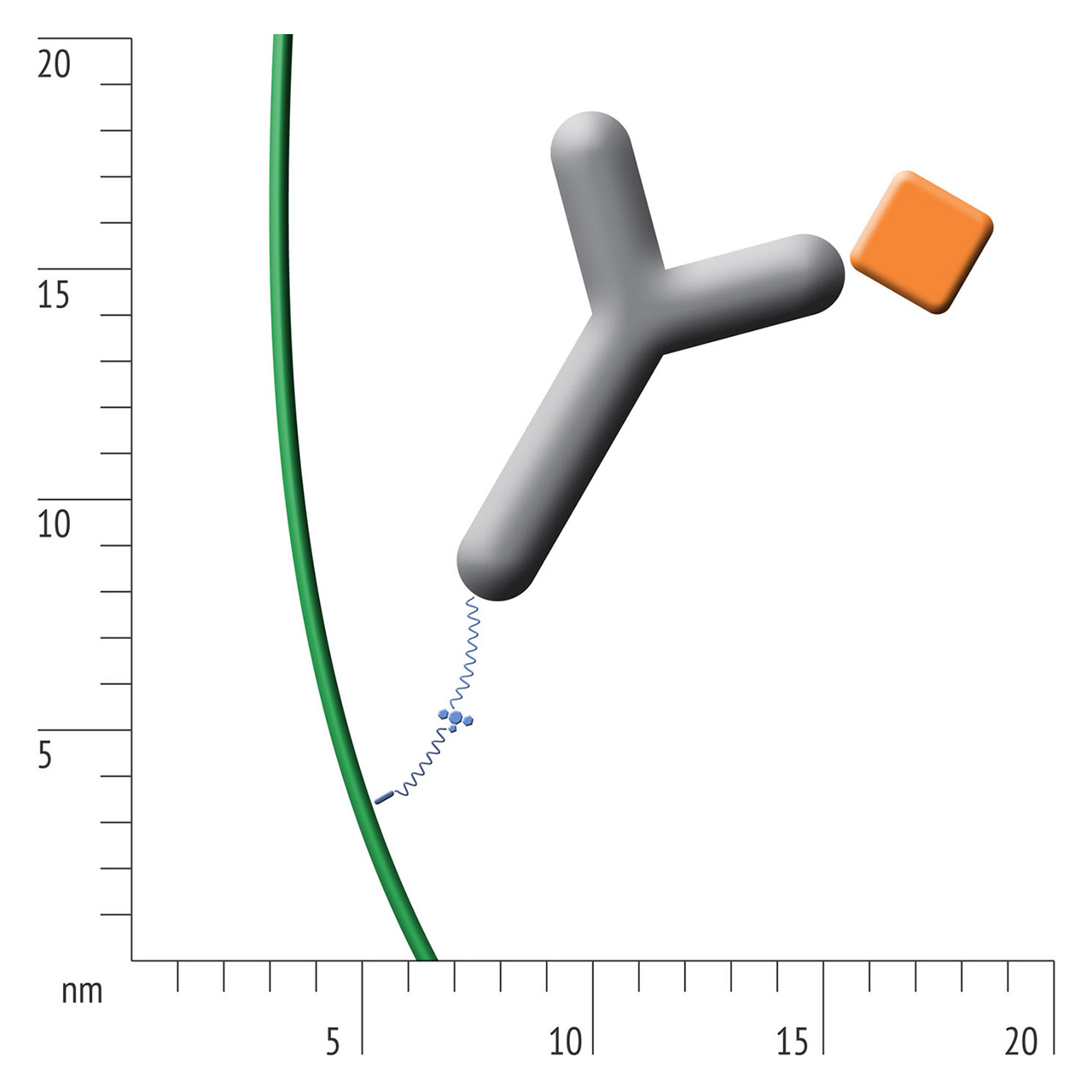
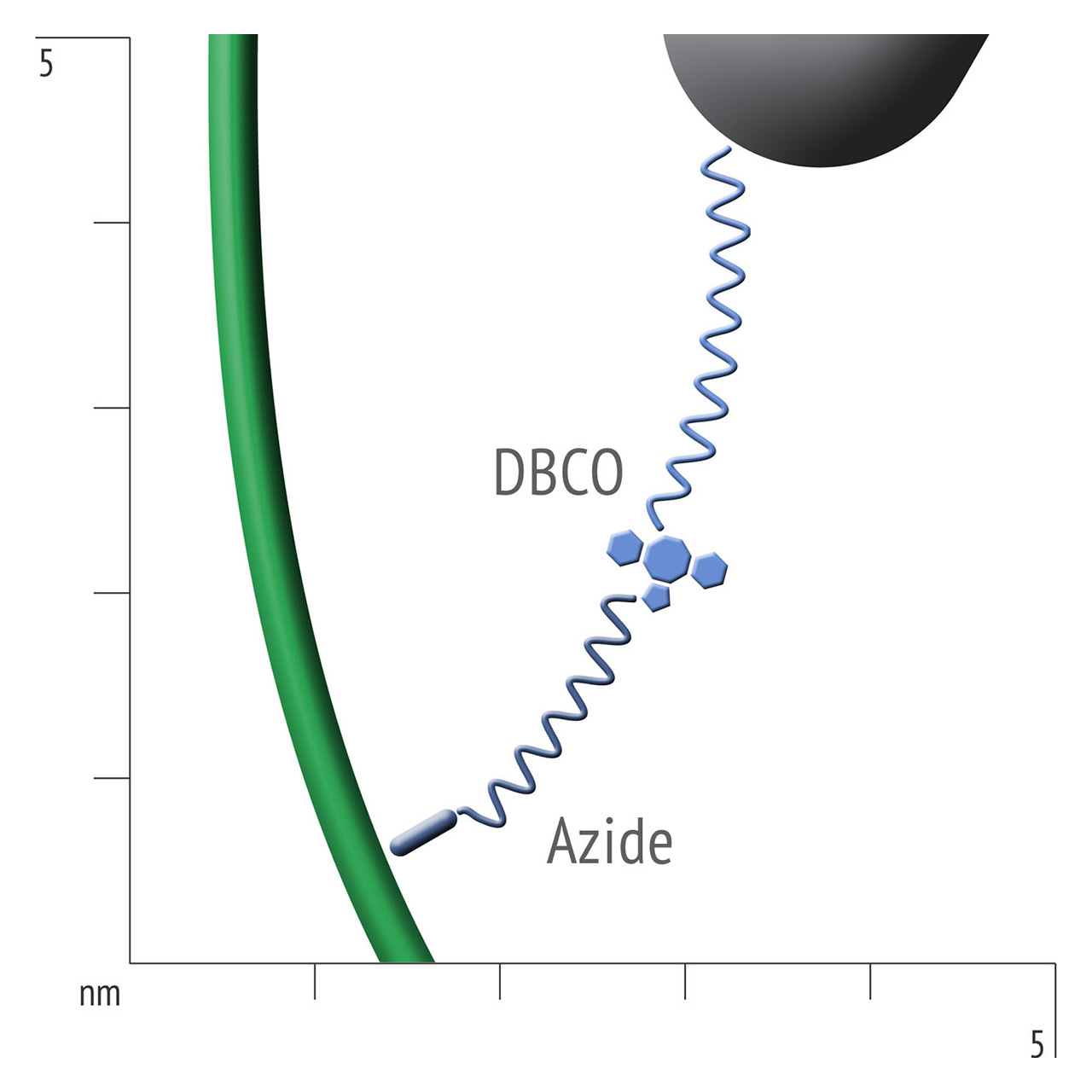
For about 20 years, Click Chemistry has gained increasing popularity among chemists, due to its outstanding reaction kinetics, excellent selectivity and superior stability in physiological environments. To make this unique bioconjugation technology available for the demanding ligand immobilization in SPR biosensing, XanTec’s experienced surface chemists modified polycarboxylate sensor coatings with a very small, bioinert azido (N3) group, while the ligand was labeled with a low molecular weight cyclooctyne linker (DBCO), a procedure comparable to ligand biotinylation. Via a simple alkyne azide cycloaddition, these groups can react in a quantitative and highly selective manner, forming a stable covalent triazole linkage.
Let’s see how your SPR experiments can benefit from Click Chemistry:
Advantages of Click Coupling
Maximum convenience:
- The sensor chip comes pre-activated, ready-to-use, and can be stored for years with practically no loss of activity over time.
- Labeling of the protein is straightforward: Add linker to the protein, incubate for a few hours, desalt, and start immobilization.
- Stored frozen, the linker is reactive for many months.
Maximum activity:
- One linker per ligand molecule maximizes ligand activity and avoids crosslinking.
- Labeling and immobilization of the ligand can be performed in physiological conditions.
Maximum control:
- The immobilization level can be adjusted at any time – even after the first interaction cycles.
- High stability and selective reactivity – no need to quench remaining reactive groups.
Maximum data quality:
- Very low non-specific binding.
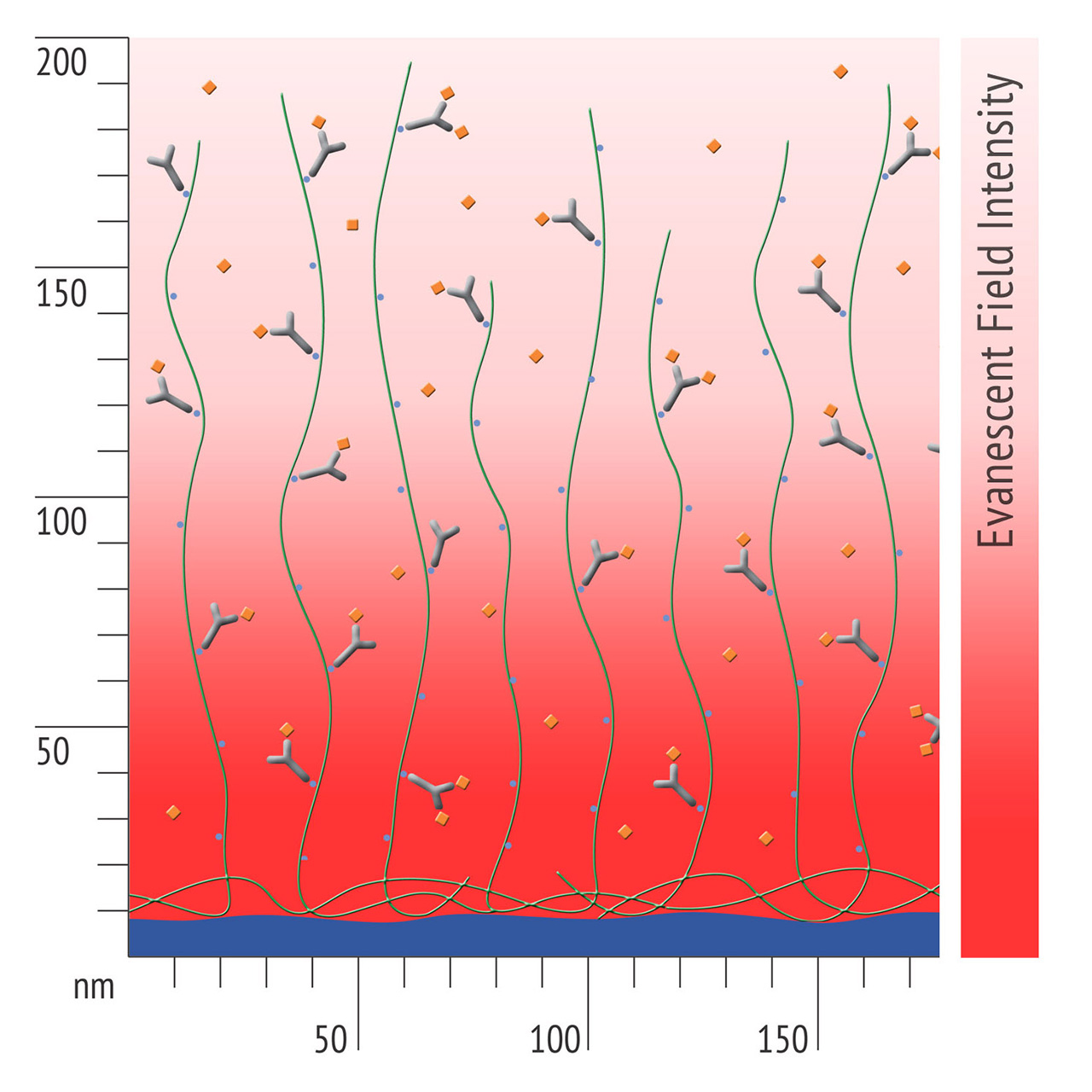
- Higher sensitivity and lower diffusion artefacts compared with streptavidin-modified sensor chips due to small linker size [Fig. 2].
Maximum flexibility:
- Use classic electrostatic preconcentration to maximize immobilization yields.
- Use physiological immobilization conditions if ligand stability is critical.
You might ask, “My SPR experiments have always worked, why should I use Click Coupling?”
Many of the disadvantages of other immobilization methods are not obvious without using sophisticated data analysis techniques, even though they negatively affect the result of an experiment and the quality of the information obtained. More obvious artifacts, such as high background, non-stochiometric interaction due to a high fraction of inactive ligand, or difficulty in controlling immobilization levels are tolerated and taken as given, although the user is well aware that the data quality could be better.
Who should use Click Coupling and why:
- Anyone who is still using EDC/NHS chemistry – to avoid crosslinking, (partial) deactivation of the ligand, and to increase the reproducibility of the immobilization level.
- Anyone who uses biotinylated ligands with streptavidin sensor chips – to avoid diffusion artefacts, to minimize non-specific binding, and to increase the immobilization yield.
- Anyone who needs reproducible immobilization levels for standardized assays to increase precision and reproducibility in QC/QA.
- Anyone whose protein tolerates only certain buffers to remain stable and active. No buffer restrictions during immobilization and analysis.
- Anyone who is struggling with high non-specific binding on NTA sensor chips.
- Anyone who intends to increase the immobilization level or add a second ligand later on, even within the actual binding experiment. Using Click Coupling, immobilization levels that are too low are a thing of the past.
Protocol for DBCO labeling
The DBCO-PEG4-NHS ester introduces dibenzylcyclooctyne (DBCO) functionality to available amine groups (in lysine residues) on a protein. The PEG4 spacer enhances solubility in water as well as in commonly used organic solvents like DMSO or DMF. Moreover, the spacer enhances the accessibility of the amine site, which may be buried within the protein.
As with every NHS ester, the DBCO-PEG4-NHS ester is moisture sensitive and hydrolyzes readily in aqueous buffers. Therefore, we recommend preparing a stock solution in dry solvent immediately before use; that is usually stable for several days. Do not use buffers containing primary amines like Tris, glycine or azides, as they will either react with the NHS ester or with the DBCO.
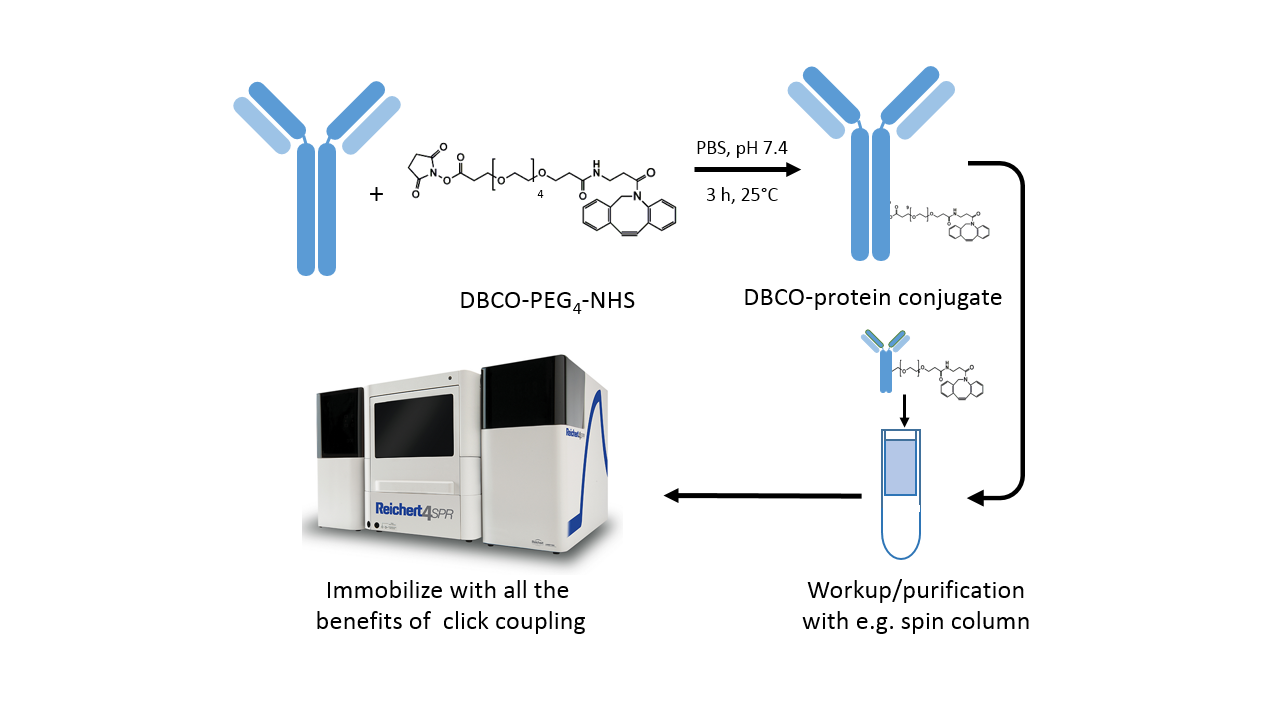
- Prepare protein solution in phosphate-buffered saline or another suitable buffer (Fig. 3).
- Immediately before use, prepare the DBCO-PEG4-NHS reagent at 10 mM in anhydrous DMSO or DMF.
- For labeling of the protein, start with a 3-fold molar excess of the labeling reagent. This prevents most proteins from being over-labeled, which causes crosslinking on the sensor chip. For lysine-poor proteins, a higher molar excess of the labeling reagent might be necessary, and vice versa for lysine-rich proteins.
- Incubate the reaction for 3 h at room temperature or 8 h on ice.
- Remove non-reacted reagent by desalting with a spin column or dialysis.
Immobilization of DBCO-labeled protein
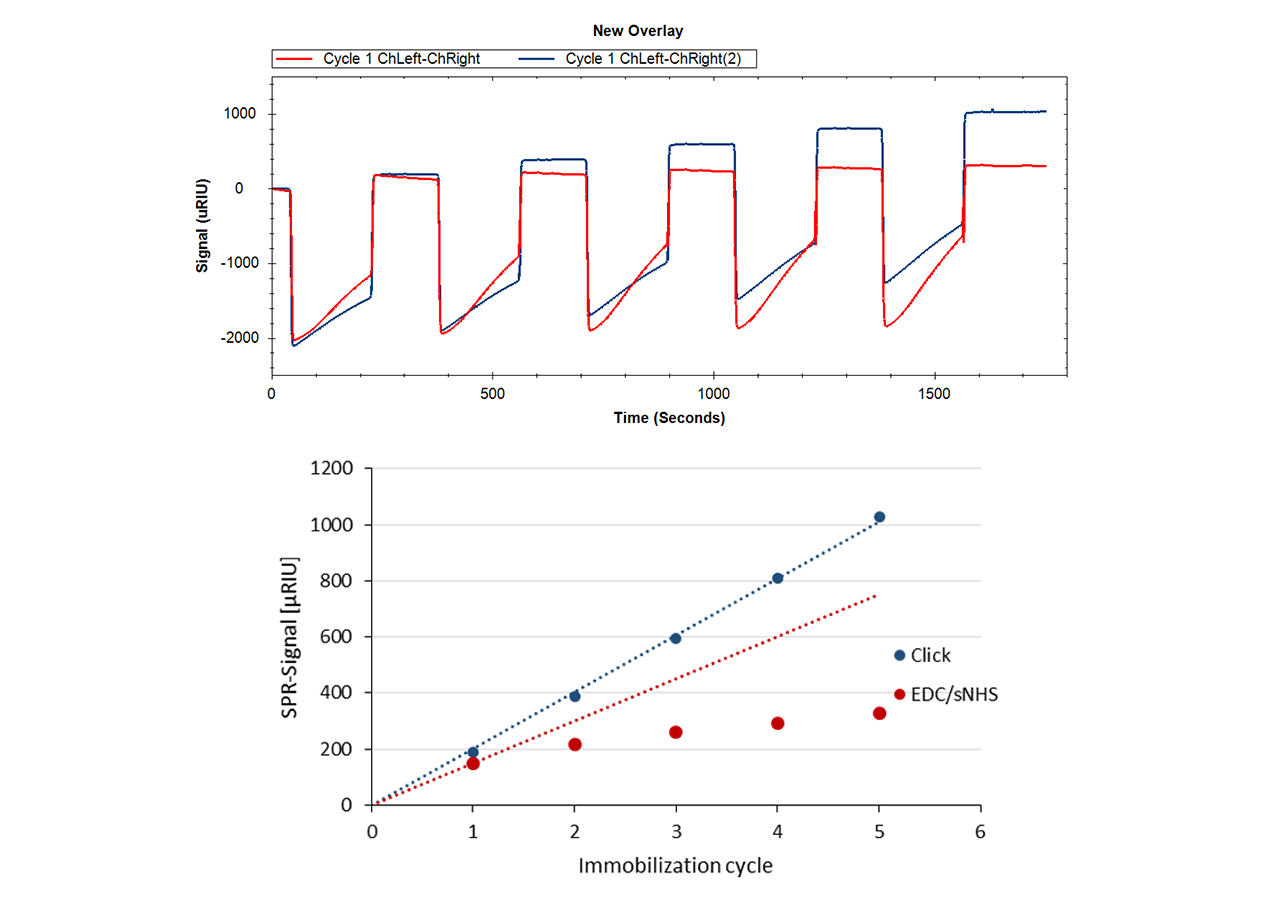
Insert an azido-sensor chip according to instructions of the instrument manufacturer. There are no restrictions on the composition of the running buffer, however added azide should be avoided during the immobilization steps. There is no need for an activation step for the azido-surface. Simply inject the DBCO-labeled protein at concentrations in the range of 10–200 µg/mL. Reaching a targeted immobilization level will require longer contact times for proteins in physiological conditions compared with electrostatic preconcentration conditions. Please keep in mind that you can always add more ligand later, as long as sufficient unreacted azido-groups are present (Fig. 4). A quenching step is not necessary, as the unreacted azido functionalities on the sensor chip are non-reactive and bioinert.
Non-specific binding (NSB)/interaction analysis/stability
Non-specific binding
Data quality in interaction analysis is influenced by many extrinsic factors. Instrument status, purity and activity of the reactants, the quality of the sensor chip, and especially the ability to reduce non-specific binding to a minimum are important for obtaining high quality data. Comparing the NSB of surfaces for traditional EDC/NHS chemistry (CMD and HC) with those that are able to bind labeled/tagged biomolecules (streptavidin/Ni-NTA/azido) reveals significant differences (Fig. 5).
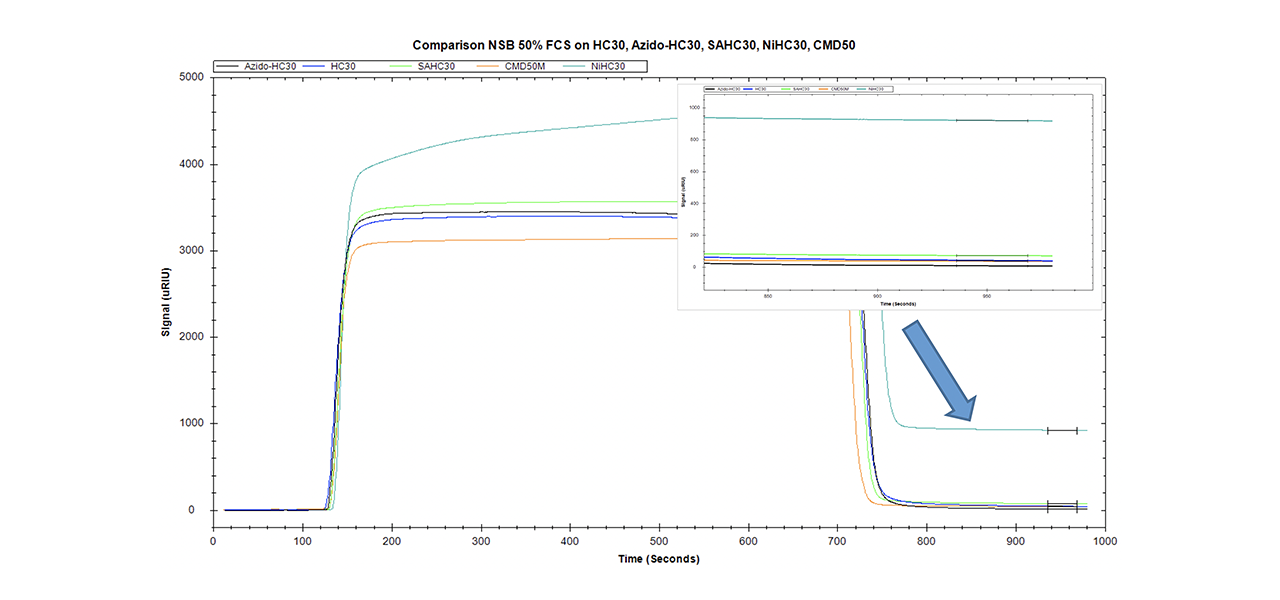
XanTec has tested the NSB of differently functionalized 30-nm linear polycarboxylate hydrogel chip coatings. In addition, a very similar sensor chip coated with 50 nm carboxymethyldextran was tested. The azido-modified polycarboxylate surface has the lowest non-specific binding of all surfaces (8.3 µRIU). Even the unmodified carboxymethyldextran (CMD, 37.5 µRIU) and linear polycarboxylate (HC, 42.1 µRIU) surfaces display a higher background. With a NSB of 72.6 µRIU, the contribution of pre-immobilized streptavidin to NSB is clearly visible from comparison with the corresponding unmodified surface. The highest NSB was for the NTA-modified chip (920.2 µRIU, NiHC30M).
Compared with the new azido derivative, non-specific binding on the streptavidin-modified surface is almost one order of magnitude higher, and on the Ni-NTA surface NSB is more than two orders of magnitude higher.
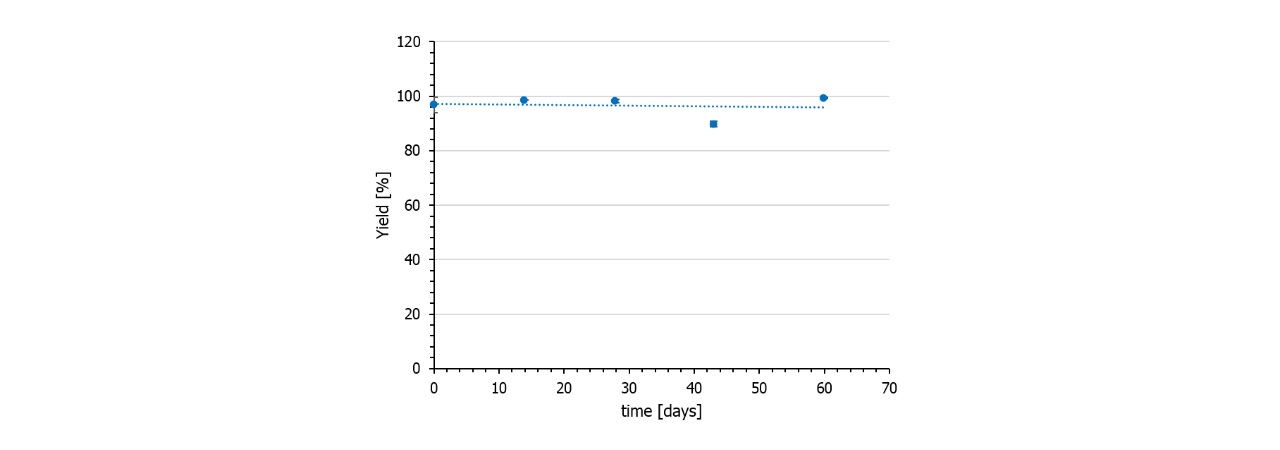
Stability/storage
In contrast to NHS preactivated sensor chips, azido surfaces are very stable and can be stored frozen for at least several months. Also, the DBCO-linker is stable for months if stored frozen. Fig. 6 shows the immobilization yield of a batch of DBCO-conjugated protein A/G on azido-functionalized HC30M sensor chips (all chips from one batch).
Interaction analysis
For interaction analysis, a well-known reference assay for SPR instruments and surfaces was chosen. Carbonic anhydrase II (CAII) was immobilized in a Reichert 4SPR system with EDC and sulfo-NHS (a more reactive ester than NHS) and with the new Click Coupling immobilization method on a plain and on an azido-modified HC30M sensor chip, respectively. The analyte sulpiride (Mw = 341 Da) in PBS was injected at six different concentrations, each in triplicate, at 25°C (Fig. 7).
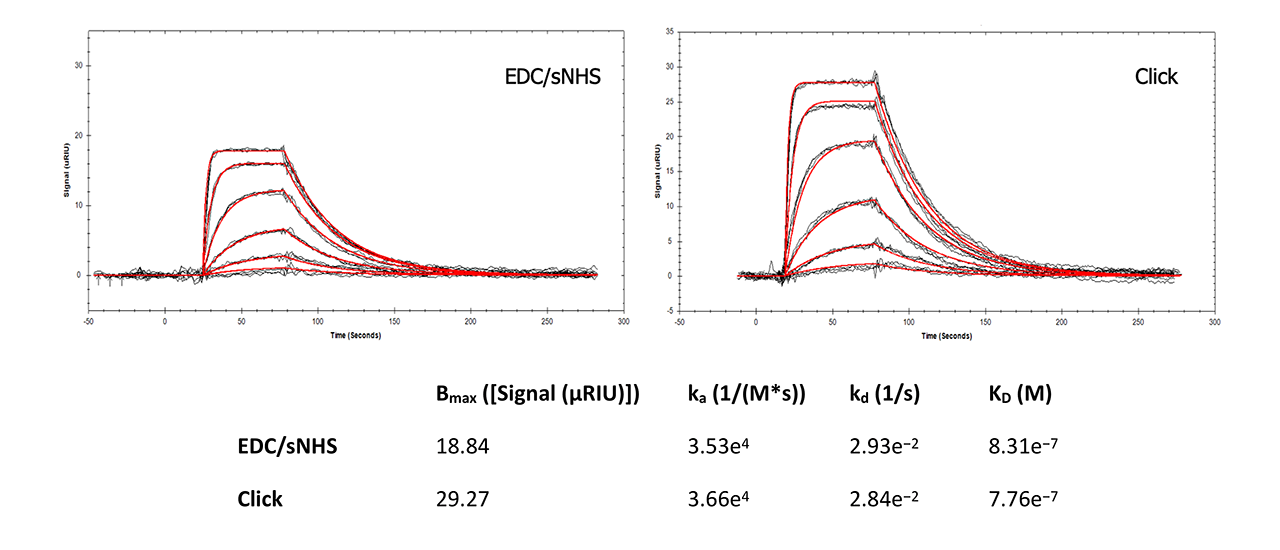
This side-by-side comparison shows that Click Coupling chemistry has no influence on the interaction of CAII and sulpiride compared with direct immobilization via EDC/NHS. All data are in good accordance with peer-reviewed, published data1, 2. Slightly higher on- and off-rates for the Click-Coupling chemistry might indicate better accessibility of the ligand by the analyte due to the presence of the PEG4 linker.
Conclusion
- Click Coupling is an efficient, universal and robust immobilization chemistry. It is particularly advantageous for sensitive proteins requiring a specific buffer system (e.g. to maintain their activity), since labeling and immobilization have virtually no buffer restrictions.
- Also, this immobilization method is very well suited for standardized assays which need to be conducted using strictly identical protocols.
- The non-specific background is significantly lower with Click Coupling compared with other chemistries.
- Other common problems, such as multiple crosslinks, scouting for optimal immobilization conditions, and difficulties reaching a sufficient immobilization level are eliminated.
- The activity of the immobilized ligand increases significantly.
- Click-Coupling allows the user to increase the ligand density step-by-step and at any time – even after the first interaction cycles.
Click-coupling is a highly repeatable, robust and ligand-friendly immobilization chemistry, which significantly increases signal-to-noise ratio and data quality.
Curious?
Surfaces for azide labelled ligands
Some expression systems are capable of site-specific integration of non-natural amino acids containing an azide moiety. For efficient, selective and site-directed Click Coupling of such ligands XanTec is also offering DBCO modified sensor chips.
References
- Navratilova, I., Papalia, G. A., Rich, R. L., Bedinger, D., Brophy, S., Condon, B., ... & Heutmekers, T. (2007). Thermodynamic benchmark study using Biacore technology. Analytical biochemistry, 364(1), 67-77.
- Papalia, G. A., Leavitt, S., Bynum, M. A., Katsamba, P. S., Wilton, R., Qiu, H., ... & Giannetti, A. M. (2006). Comparative analysis of 10 small molecules binding to carbonic anhydrase II by different investigators using Biacore technology. Analytical biochemistry, 359(1), 94-105.