NTA–modified sensor chips (Ni)
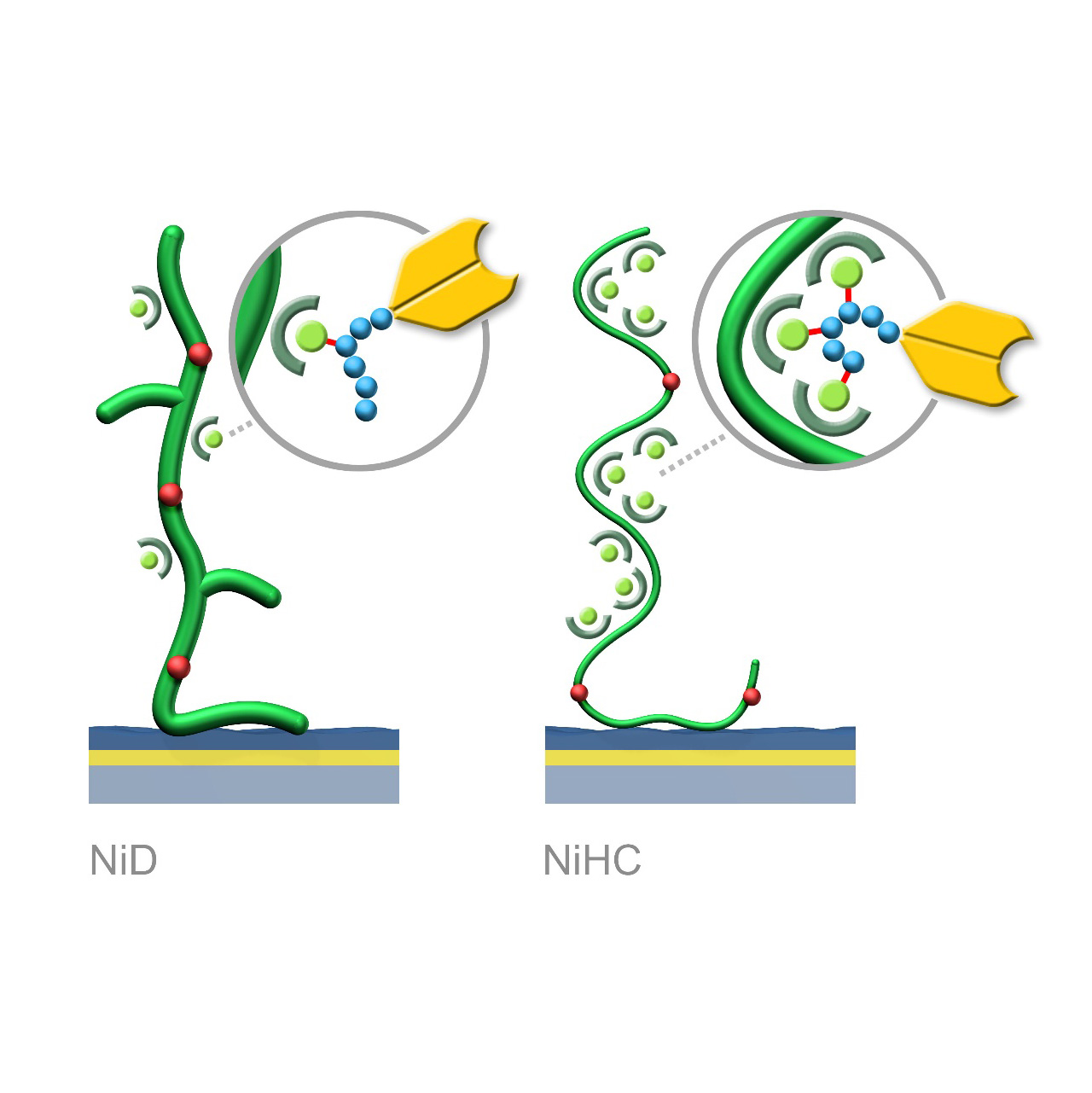
XanTec’s NiD and NiHC sensor chips are coated with a bioinert (poly)carboxylate hydrogel matrix modified with nitrilotriacetic acid (NTA) functionalities. These sensor chips enable the reversible capture immobilization of His-tagged biomolecules through complex formation with transition metal ions, preferably Ni2+. For sufficient binding stability, the His-tag should contain 6–10 histidine residues. Complete regeneration of both NiD and NiHC surfaces is typically achieved by removal of chelated Ni2+ ions using chelating agents (e.g., EDTA) or competitive ligands such as imidazole.
Binding characteristics:
NiD sensor chips exhibit predominantly monovalent interactions between NTA(Ni2+) complexes and His-tags, resulting in dissociation rates (koff) in the range of 10−3 s−1.
NiHC sensor chips support multivalent binding due to the flexibility of the polymer chains, increasing binding stability by up to three orders of magnitude. Baselines after His-tagged ligand capture show minimal to no drift, with typical koff values of 10−5–10−6 s−1.1
A known limitation of NTA-based capture surfaces is the potential for nonspecific interactions between chelated Ni2+ ions and protein analytes or complex sample matrices (e.g., FBS). Proteins containing exposed histidines or metal-affinity motifs may bind non-selectively, increasing background signals. For assays sensitive to nonspecific binding, alternative immobilization strategies (e.g., anti-His-tag antibody capture) should be considered.
Key features:
- Fast assay development: No preconcentration or surface activation required; reversible ligand immobilization under physiological conditions.
- Easy regeneration: Chip surfaces regenerated using EDTA or imidazole injections.
- Outstanding stability (NiHC): Multivalent binding minimizes baseline drift during analysis.
- Oriented immobilization: His-tag–directed capture ensures maximum ligand activity.
- Versatile capture capacity: Available on CMD and HC base coatings for a broad range of analytes.
| Product code | NiP | NiD200M | NiHC200M | NiHC1500M |
|---|---|---|---|---|
| Base coating | 2D, ultra-short bioinert CM-dextran (high density) |
3D, 200 nm bioinert CM-dextran (medium density) |
3D, 200 nm bioinert polycarboxylate (medium density) |
3D, 1500 nm bioinert polycarboxylate (medium density) |
| Capture immobilization capacity [µRIU]4 | ≈ 120 | ≈ 400 | ≈ 1,200 | ≈ 2,000 |
| Ligands | His-tagged peptides and proteins | |||
| Recommended analytes |
|
|
|
|
| Intended purpose |
|
|
|
|
1 Maximum binding stability requires immobilization at approximately one-third of the chip’s maximum capture capacity.
2 All illustrations are schematic representations and are not drawn to scale; dimensions, densities, and spatial relationships do not reflect actual physical or chemical proportions.
3 Table includes a selection from XanTec’s full Ni sensor chip portfolio.
4 Binding capacities are based on the capture level of His6-peptide from a 5 µM solution after a 180 s stabilization period, with 1 µRIU corresponding approximately to 1 RU. Immobilization capacities of His-tagged proteins are considerably higher.