XanTec bioanalytics specializes in manufacturing high quality sensor chips compatible with all major surface plasmon resonance (SPR) instrument brands on the market. Through consistent research, XanTec has developed the widest portfolio of sensor chips available today, offering tailor-made solutions for almost any application.
Fragment-based drug discovery (FBDD) has become a preferred alternative to high-throughput screening (HTS) to improve the discovery of small-molecule drug candidates. Screening of low-molecular-weight fragments can identify hit compounds with better efficiency and physiological profiles than HTS2.
SPR biosensor technology is one of the primary biophysical methods to screen fragment libraries3 as newer instruments achieve sufficiently high signal-to-noise ratios to generate reliable data despite the low molecular weight and low affinity of many analytes.
In previous approaches to establish FBDD assays using SPR, the ligand was covalently immobilized on the sensor surface with high immobilization levels to ensure that the protein bound stably to the sensor chip surface. Alternatively, biotinylated proteins were immobilized on streptavidin coated sensor surfaces with the inherent drawback that the analyte could non-specifically interact with the streptavidin. Both immobilization methods lack the possibility to remove the bound ligand from the sensor surface which is critical, for example, when working with GPCRs or other sensitive proteins which often denature over long screening campaigns.
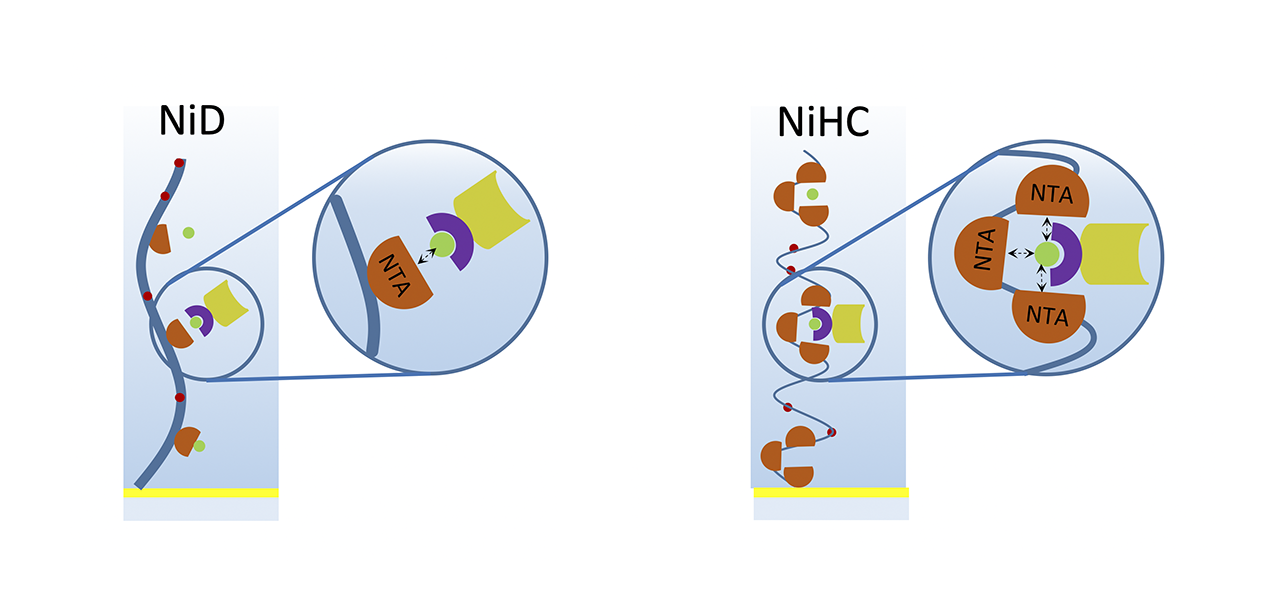
To make this group of sensitive molecules accessible to FBDD, various attempts have been made to immobilize them reversibly via affinity based His6/nickel-nitrilotriacetic acid (NTA) coupling (Fig. 1). State-of-the-art NTA sensor chips are based on carboxymethyl dextran (CMD) hydrogel modified with NTA groups. Because of the relatively low affinity between the His6-tag and the Ni2+-NTA-complex, continuous dissociation of the ligand from the sensor chip surface leads to unwanted baseline drifts. Such drift effects can easily exceed the specific signal when screening small molecules and thus represent a major problem.
To account for this well-known disadvantage, surface chemists at XanTec developed a poly-NTA sensor chip hydrogel coating based on a strongly hydrated and very flexible polycarboxylate polymer backbone. Compared to the standard CMD-NTA chemistry, these new coatings, which are available in 30, 200, 1000 and 1500 nm thickness, can improve the stability of captured His6-tagged ligands by 2-3 magnitudes, matching the high affinity of the recently-developed Tris-NTA4.
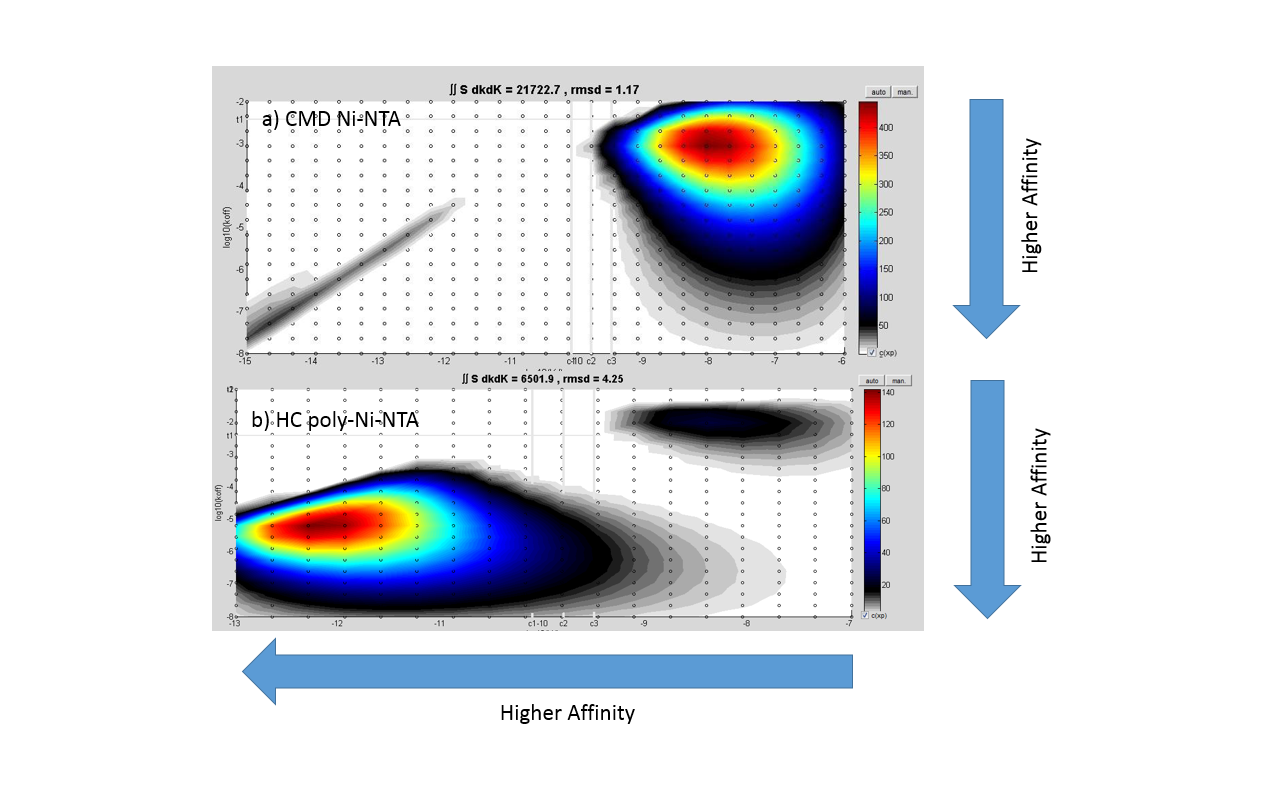
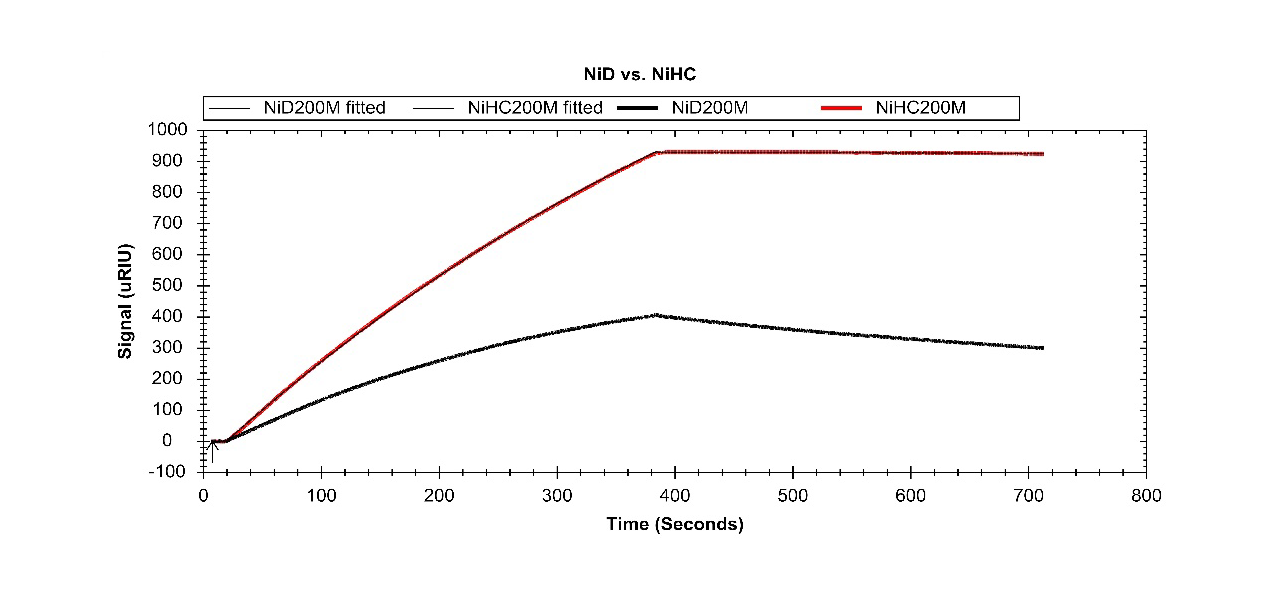
Figures 2 and 3 are showing the much higher stability of a captured His6-tagged ligand (a protein A/G fusion protein) on XanTec’s poly-NTA surface NiHC1000M than on NTA-derivatized CMD hydrogel (mono-NTA). Despite the high affinity of this surface, the regeneration conditions (EDTA) are mild, and identical to those of the standard mono-NTA sensor chips.
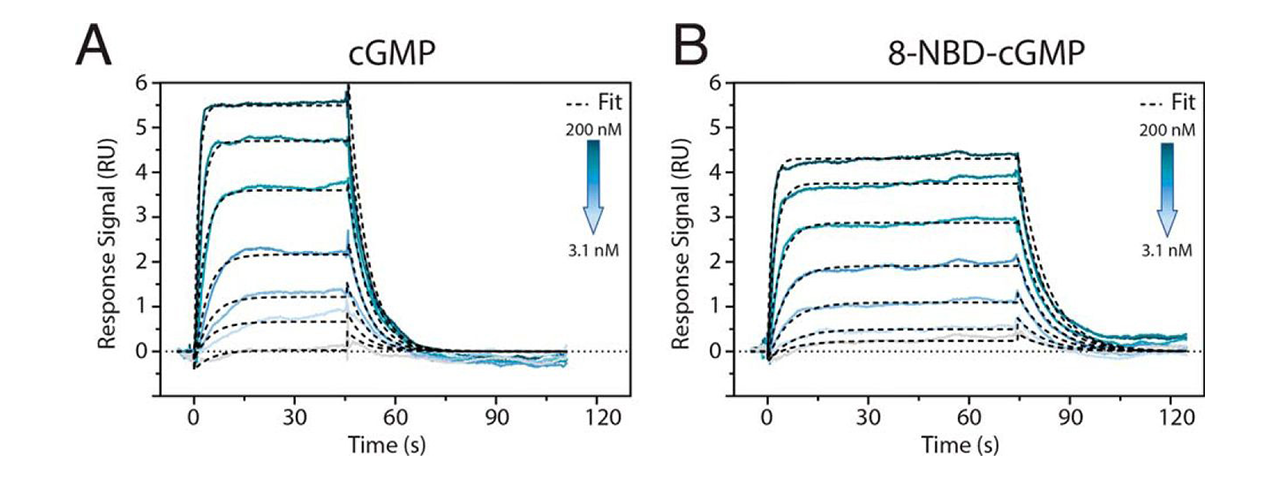
The effective use of polydentate high-affinity Ni-NTA surfaces in SPR measurements of small molecules is vividly demonstrated in a publication by Byun et al.6. A direct binding assay was used to investigate the kinetics of cGMP (345 Da) and a cGMP analog (8-NBD-cGMP, 605 Da) binding to the cGMP-dependent protein kinase of Plasmodium falciparum (PfPKG), the malaria-associated parasite. Even at high immobilization capacities, the XanTec bioanalytics high-affinity NiHC NTA sensor chip used showed no leaching effects, so Rmax values well below 10 RU could be evaluated (Fig. 4).
Conclusion
With XanTec’s unique poly-NTA sensor chips (NiHC group) it is possible to establish higher immobilization levels compared to NTA-derivatized carboxymethyl dextran with the additional benefit of drastically reduced leaching, resulting in a practically drift-free baseline. This allows repeated immobilization of sensitive ligands during extended FBDD campaigns, as NiHC chip surfaces are fully regenerable over many interaction cycles.
References
- Faoro, C., Wilkinson-White, L., Kwan, A. H., & Ataide, S. F. (2018). Discovery of fragments that target key interactions in the signal recognition particle (SRP) as potential leads for a new class of antibiotics. PloS One, 13(7), e0200387.
- Retra, K., Irth, H., & van Muijlwijk-Koezen, J. E. (2010). Surface Plasmon Resonance biosensor analysis as a useful tool in FBDD. Drug Discovery Today: Technologies, 7(3), e181-e187.
- Shepherd, C. A., Hopkins, A. L., & Navratilova, I. (2014). Fragment screening by SPR and advanced application to GPCRs. Progress in Biophysics and Molecular Biology, 116(2-3), 113-123.
- Lata, S., Reichel, A., Brock, R., Tampé, R., & Piehler, J. (2005). High-affinity adaptors for switchable recognition of histidine-tagged proteins. Journal of the American Chemical Society, 127(29), 10205-10215.
- Gorshkova, I. I., Svitel, J., Razjouyan, F., & Schuck, P. (2008). Bayesian analysis of heterogeneity in the distribution of binding properties of immobilized surface sites. Langmuir, 24(20), 11577-11586.
- Byun, J. A., Van, K., Huang, J., Henning, P., Franz, E., Akimoto, M., ... & Melacini, G. (2020). Mechanism of allosteric inhibition in the Plasmodium falciparum cGMP-dependent protein kinase. Journal of Biological Chemistry, 295(25), 8480–8491.
Contact us today to make the most out of your assay and optimize your data. Our application specialists are available to answer any technical questions and will happily discuss your requirements.