Surface Plasmon Resonance (SPR) is an invaluable technique that generates information-rich data for a variety of biomolecular interactions. Researchers use SPR to understand biological pathways and to develop and characterize a range of potential therapeutics to treat disease. These interactions include those occurring with and between the major classes of biological macromolecules.
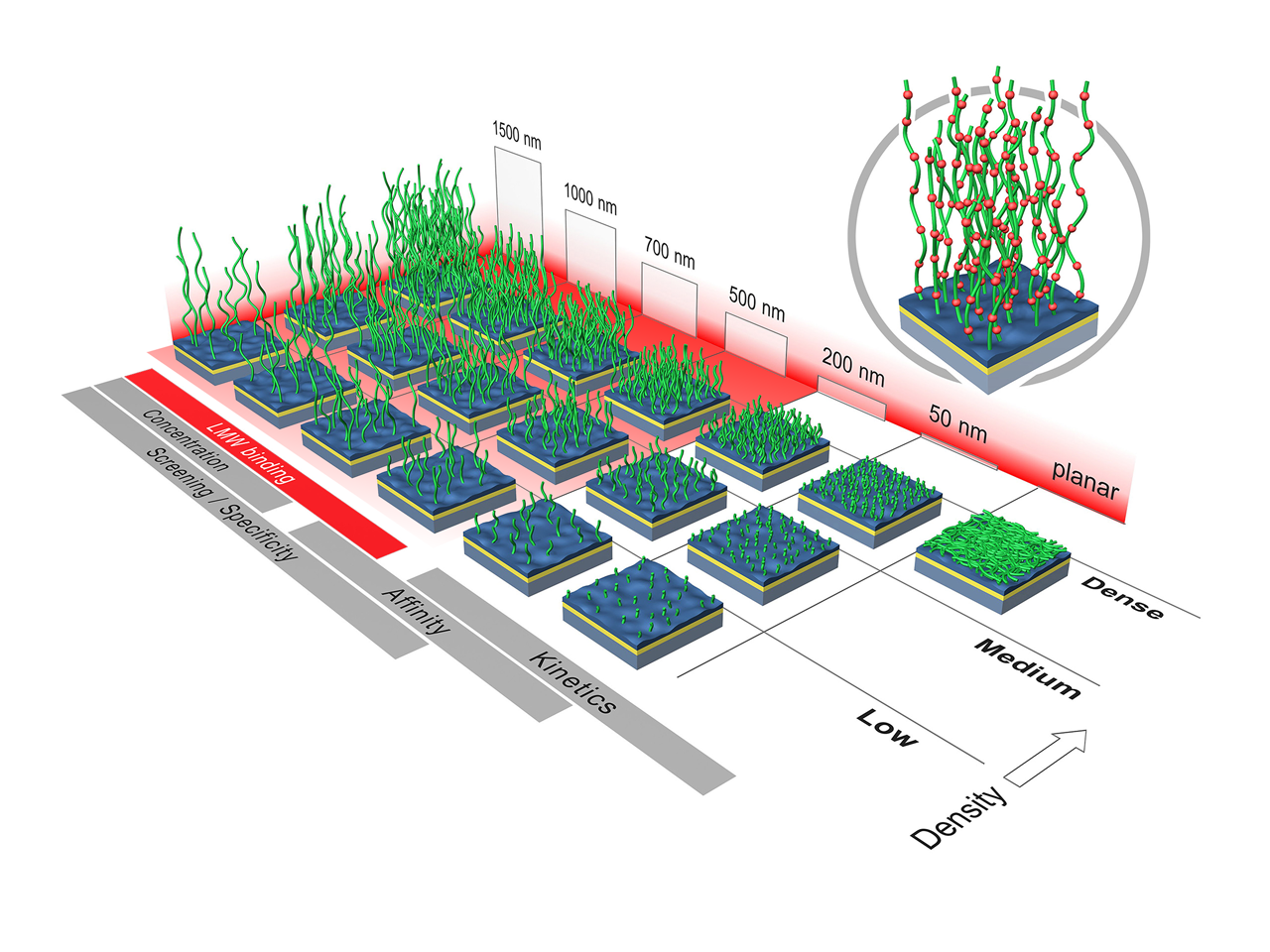
This article is focused on using SPR to study biomolecular interactions involving small molecules binding to proteins. Many researchers rely on SPR to provide key information about these interactions; the technique is widely used for the determination of kinetic and thermodynamic parameters. For the purposes of this article, “small molecules” generally refers to molecules with molecular weight ≤1,000 Da. There are certain standard approaches that researchers follow to carry out these types of experiment. As a higher surface density is needed when there is a large molecular weight difference between the protein (the target/ligand) and the small molecule (the analyte) whose interaction is being studied, direct coupling is usually the preferred means of attaching a target to the sensor chip.
XanTec bioanalytics offer a broad range of sensor chips with high immobilization capacities for small molecule screening. Depending on the actual size of the ligand and analyte, there are different thicknesses, densities and even different polymer types to choose from. Thus, the results of screening campaigns or kinetic/thermodynamic analyses can be optimized. Several high capacity coatings are also available as derivatives, such as with NeutrAvidin/streptavidin, or (poly)NTA for His-tagged ligands.
Since it is common for small molecules to show limited solubility in the aqueous running buffers used for SPR studies (eg. PBS, HEPES), analytes soluble in DMSO are typically diluted with running buffer to about 1-5% DMSO prior to SPR analysis. In some cases, detergents may also be used to help with solubility issues. Many of these experiments are carried out to aid in the development of small molecules as potential therapeutics for disease. We now highlight some recent articles that researchers have published using Reichert’s SPR systems for investigating low molecular weight molecules interactions with proteins.
Example 1: Target HIV – using a Reichert4SPR for screening
A 2019 article in ACS Infectious Diseases highlights how some of the unwanted functions of HIV Nef accessory protein, which plays a key role in HIV replication, can be blocked by small molecules1.
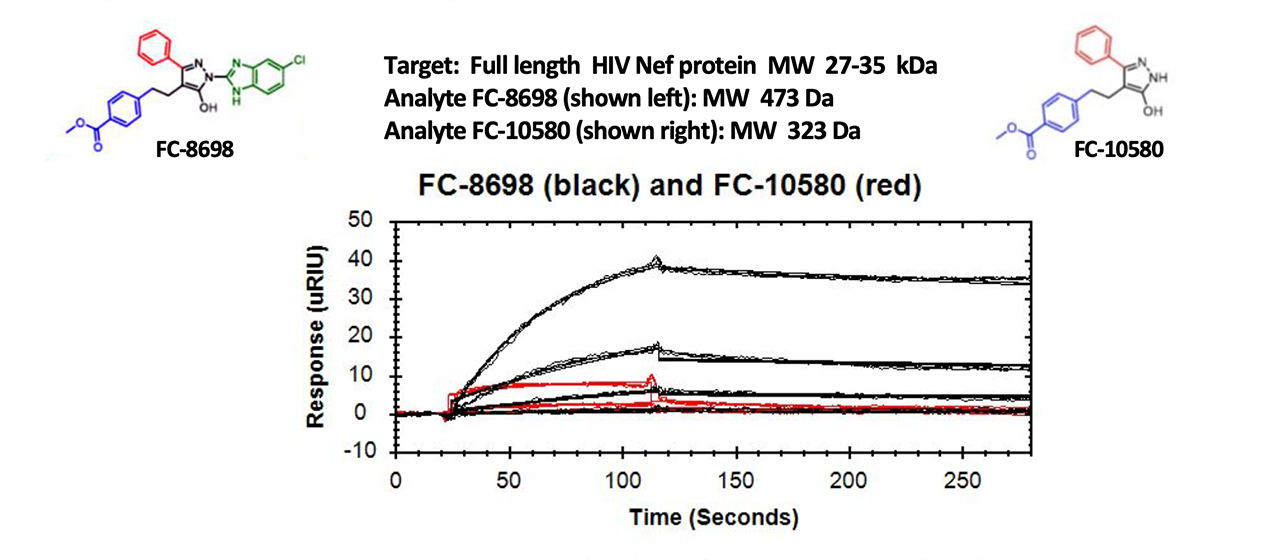
For the results presented in this article, researchers developed analogs to an earlier small molecule that showed promise in testing. They had more than 200 analogs to analyze and ranked them based on binding results obtained using a Reichert4SPR system. Recombinant HIV-1 Nef was covalently bound to a dextran sensor chip as the target and binding interactions with the analogs were ranked in terms of affinity, residence time and binding response1. Of the 216 analogs screened, 45 exhibited no binding. The remainder of the responses were ranked based on the magnitude of the responses, their kinetics, including on- and off- rates, and activity from a separate assay1.
SPR results for the two compounds shown below are representative of how the researchers determined that the benzimidazole moiety at position C is essential for Nef inhibitor action. As noted in the article, the percent inhibition of HIV-1 infectivity determined using the TZM-bl reporter cell assay goes down by nearly 90% if the benzimidazole moiety at position C (highlighted in green below) is not present on the scaffold. In addition, the affinity of the interaction decreases significantly from 13 nM to 9.8 μM1 and the absolute magnitude of the responses decreases approximately fourfold (see overlays below). Based on these results, FC-8698 was ranked as one of the top overall binders of the compounds studied.1
Example 2: Target Tuberculosis – using a two-channel Reichert SPR for initial screening
A 2017 article in ChemMedChem describes how an SR7500DC (2-channel Reichert SPR instrument) in combination with a high capacity streptavidin sensor chip from XanTec bioanalytics (SAD500M) was used for the screening of potential antimycobacterial agents against Mycobacterium tuberculosis2.
In this article, researchers explain that there is an urgency to their work because multi-drug resistance and the need for long term treatment are making it difficult to treat this disease and, they note that there are millions of new cases of Mtb infections each year worldwide (as of the year 2017 when they wrote their article)2. Previous work showed that the enzyme CYP121 is essential for Mtb growth making it a potential target for Mtb treatment.2
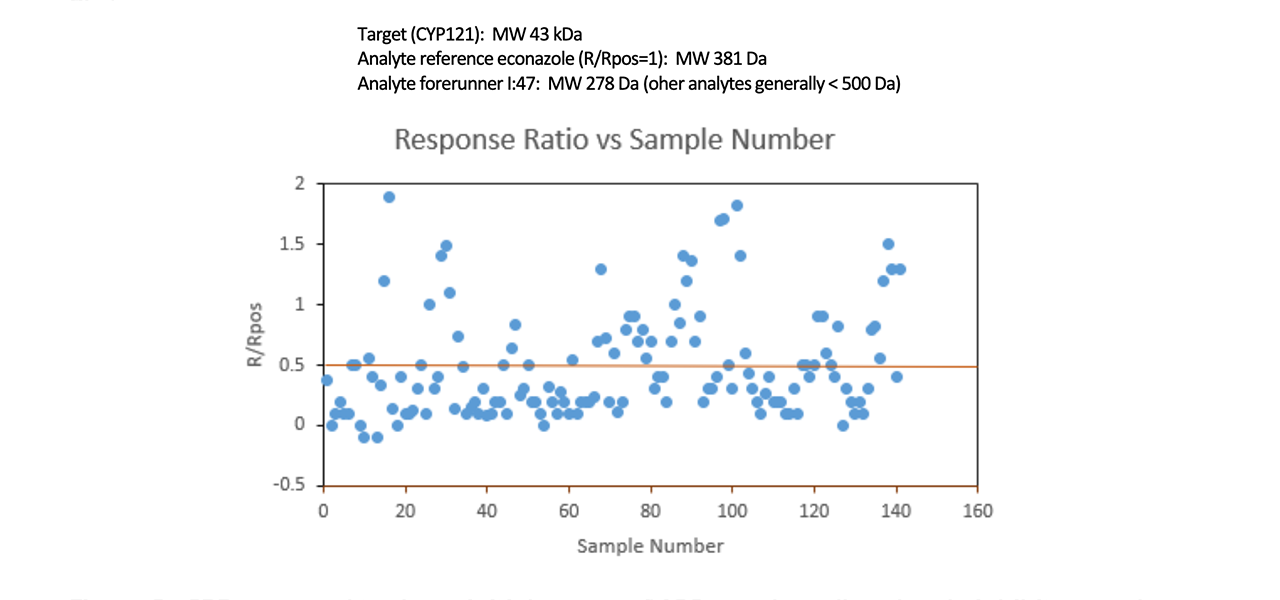
Screening in this study was carried out based on biophysical and microbiological methods. The initial screen was carried out using Surface Plasmon Resonance (SPR) with a Reichert 2-channel instrument and involved capturing biotinylated CYP121 on a streptavidin chip as the target and then analyzing binding to the target of 139 novel small molecule antimycobacterial compounds across six classes. Researchers built up small molecule compounds on six different scaffolds to probe a wide range of structures. Of the compounds screened, 32% showed a response using SPR. Seventeen of the compounds had higher responses than econazole, the positive control.
When compared to the positive control econazole, forty-four of the compounds screened were found to be worthwhile for further testing (ie. to have responses R/Rpos of 0.5 or higher). Further testing with multiple techniques led the researchers to a promising target for further research, a compound they call I:47 which exhibited good antimycobacterial activity and bacterial selectivity with a favorable toxicity profile2.
Example 3: Anti-Cancer Drug Candidates – comparison using a Reichert4SPR
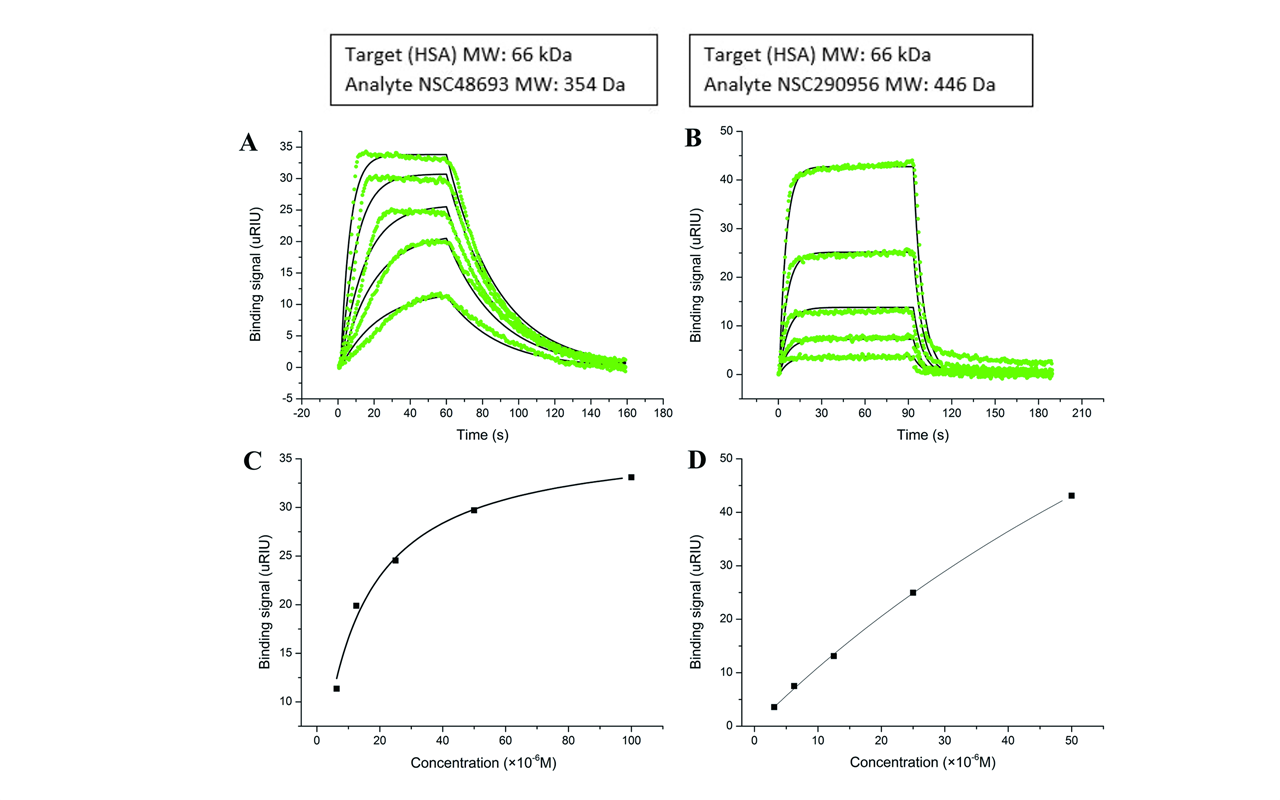
A 2017 Article in PLOS 1 includes SPR data obtained using a Reichert4SPR for the binding of small molecule anti-cancer drugs to human serum albumin (HSA)3. In this study, researchers looked at two potential anti-cancer drug candidates, NSC48693 (which is hydrophilic) and NSC290956 (which is hydrophobic) and determined where they bind to HSA using fluorescence quenching and molecular modeling. For information regarding the kinetics and thermodynamics of the small molecule/HSA interactions, researchers used a Reichert4SPR.
HSA was amine coupled to a dextran sensor chip and then the two drug candidates were diluted and injected over the surface as analytes. Kinetic analysis resulted in the determination of the affinity for the HSA/NSC48693 interaction (KD = 13.8 μM) to be almost 10 times higher than the affinity for the HSA/NSC290956 interaction (KD = 116 μM).3
This information is expected to be particularly useful in later determination of the type of dosing that would be needed for clinical studies using each compound.
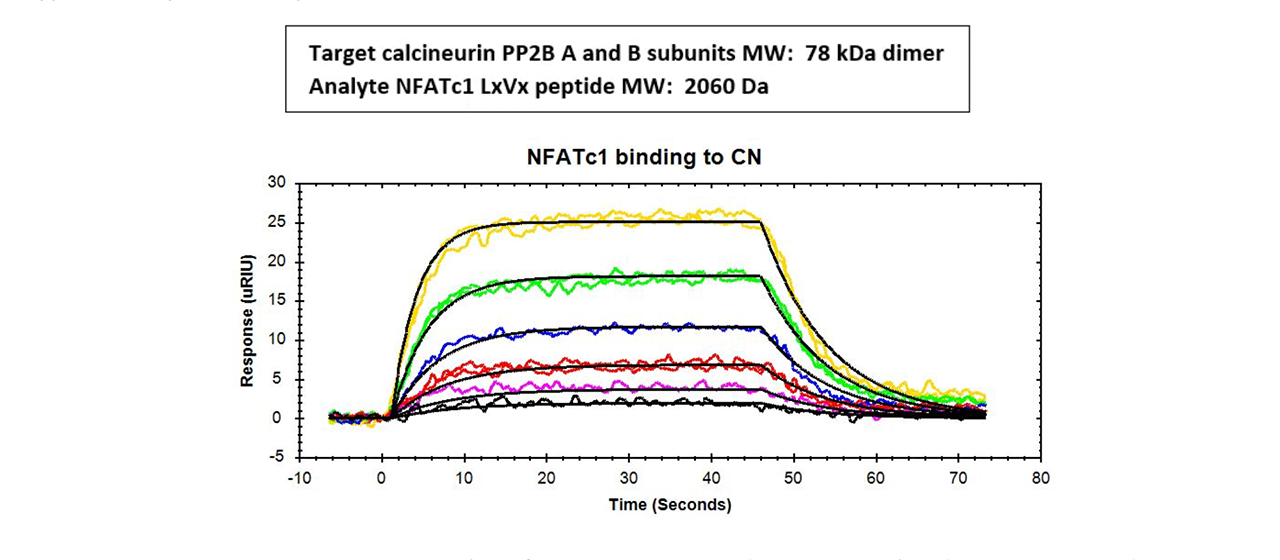
Example 4: The Binding of short linear motifs (SLiMs) to the Enzyme Calcineurin
In a 2019 paper in ACS Chemical Biology researchers present results from their research into two known short linear motifs (SLiMs), PxIxIT and LxVP, which bind to Calcineurin (CN), an enzyme involved in a number of biological functions including such things as development, heart function, and immune response4. Although most cellular signaling involves protein/protein interactions, some also occurs between proteins and short linear motifs which are about 3-10 amino acids in size and are typically located in intrinsically disordered regions. Using SPR to investigate these interactions is not trivial because the motifs are small and have low binding affinities.
For this project, researchers captured His-tagged CN on a NiNTA sensor chip and used a Reichert4SPR surface plasmon resonance (SPR) instrument to determine binding constants with peptides containing either PxIxIT or LxVP. Researchers employed Structure Based Shape Complementarity (SBSC) analysis which uses the shape of the preformed, rigid SLiM binding pocket to identify complementary amino acids for binding to expand the number of known SLiM sequences. Using this approach, they identified 119 CN regulators and substrates containing either PxIxIT or LxVP. Even though this type of interaction is usually weak, they were able to obtain kinetics. From SPR results, researchers concluded that LxVP SLiM binding to CN is on-rate-dependent4.
Results for a typical compound they studied are shown here:
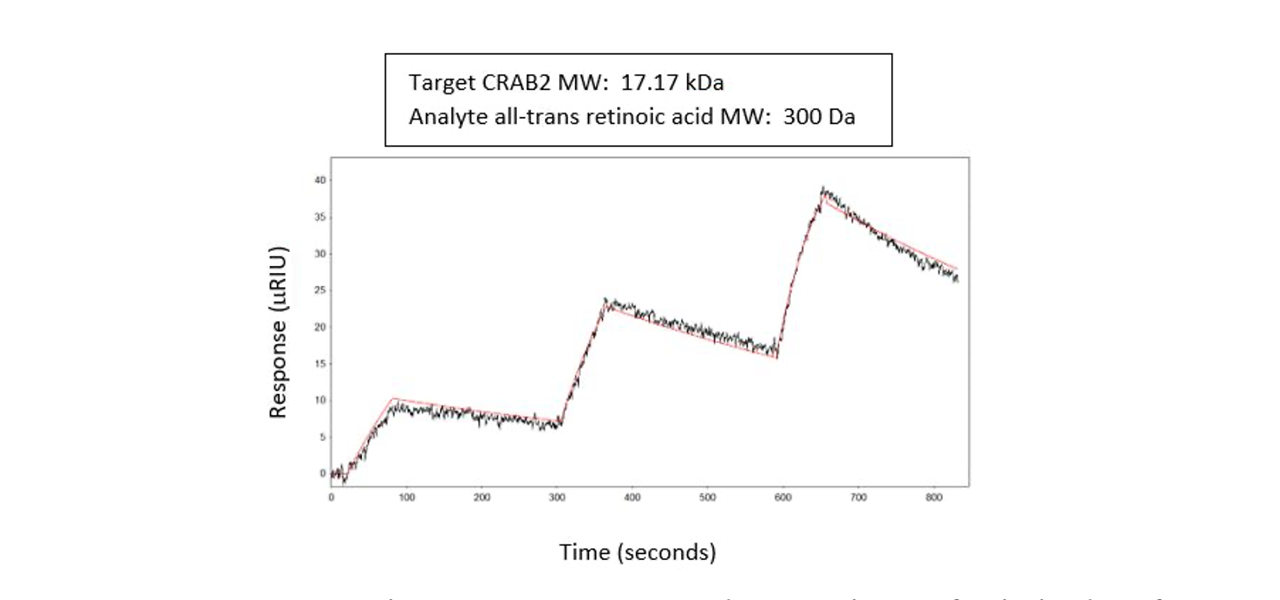
Example 5: Retinoic Acid – an important nutrient
In a 2019 article in Biochemistry, research on the binding of all trans Retinoic Acid (atRA), a metabolite of Vitamin A, to Cellular retinoic acid-binding protein 2 (CRABP2), an intracellular lipid- binding protein, is described5. CRABP2 delivers atRA to Retinoic Acid Receptors (RARs), which than leads to the activation of specific gene transcription. Using a Reichert4SPR, they captured His- tagged CRABP2 on a Ni-NTA sensor chip and injected the atRA over as analyte.5 All-trans retinoic acid (atRA) is a hydrophobic molecule and CRABP2 controls its movement between the nucleus and cytoplasm.
They determined that the affinity for the CRAB2/atRNA interaction was in the nM range with a slow off rate. This information, combined with the hydrophobic nature of CRABP2, indicated to them that the atRA release from the CRABP2 binding pocket was probably mediated by external factors. They noted that the binding rate was determined by atRA’s rate of diffusion out of detergent, which had to be added to the solution due to atRA’s poor aqueous solubility5.
A typical result is shown here:
Example 6: Development of CK2 inhibitors for Cancer Treatment
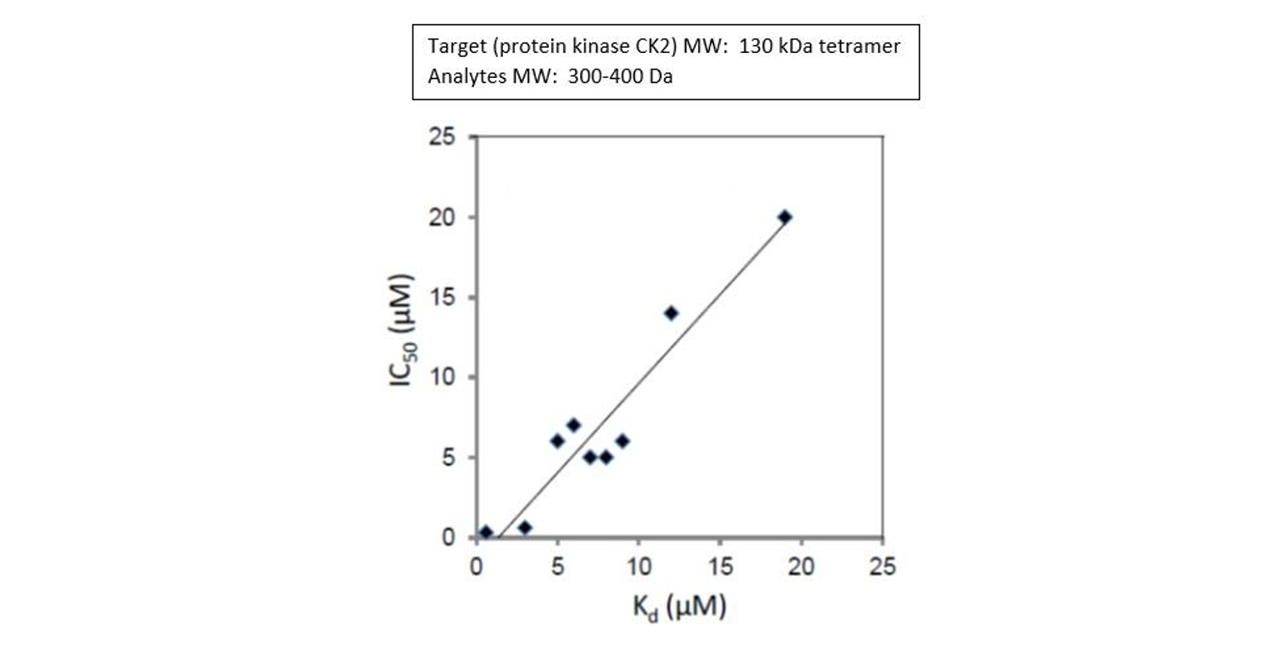
In a 2019 article in the Journal of Medicinal Chemistry, researchers used the protein kinase CK2 as target and looked for potential inhibitors6. The protein kinase CK2 is known to play an important role in pro-oncogenic pathways that are critical to cells including proliferation, differentiation, and survival. Previous research involving development of inhibitors for the protein kinase CK2 were targeted toward the ATP binding pocket, but it was found that the compounds developed could also act on other targets in the body with ATP binding pockets which could then cause unwanted side effects. Researchers in this study previously determined there was a more specific, potentially druggable site on the protein CK2. In their current work they sought to prove it to be a viable option by developing 2-Aminothiazole Derivatives that they could test.
Researchers amine coupled Anti- GST to the surface of a dextran sensor chip and then captured GST-tagged CK2α over the surface as a target for the allosteric inhibitors they made6. Of the allosteric inhibitors they constructed and tested, they found one of the compounds, 2-hydroxy-4-((4-(naphthalen-2-yl)thiazol-2- yl)amino)benzoic acid to be the lead compound with a sub-micromolar IC50. Testing was carried out using a number of techniques including SPR6. Comparing IC50 (biochemical inhibition) results with SPR binding affinity constants obtained with a 2-channel Reichert SPR for a number of the inhibitors they tested, they found good agreement7.
Example 7: Potential Anti-Influenza Agent
In a 2016 paper in Scientific Reports, researchers describe how using a “wedge” could destabilize matrix protein 1 (M1) of influenza A virus (IAV) to and keep it from self-associating8. They started out testing about 70,000 potential compounds using virtual screening. Then, they took the top ten hits and did in vitro testing and found that six of the compounds showed antiviral activity. However, five of the six also exhibited substantial cytotoxicity. One small molecule which they call PHE only showed minimal cytotoxicity and became their top candidate to carry into further testing8. PHE is a hydrophobic compound with a molecular weight 400 Da and was found to be straightforward to synthesize in just a few steps so it was carried forward into further “proof of concept” testing. Researchers used a 2-channel Reichert SPR to determine kinetics for M1 to itself and M1 to PHE. Biotinylated M1 was captured over a neutravidin surface. M1 was then injected over at various concentrations and affinity was determined (Figure 7).
Next, they tested M1 binding to PHE and determined the affinity (Figure 7). The slow off rate for the M1/M1 interaction led researchers to conclude that self-association is an off-rate driven process8. The affinity decreases from about 50 pM for M1 to M1 binding to 870 nM for PHE binding to M1 which researchers stated is still fairly tight binding for a small molecule. Using molecular modeling, they found that PHE binds to multiple sites on M1 which they feel may give it a better chance of destabilizing M1. The method of inhibition is that the small molecule impedes M1 oligomerization.8

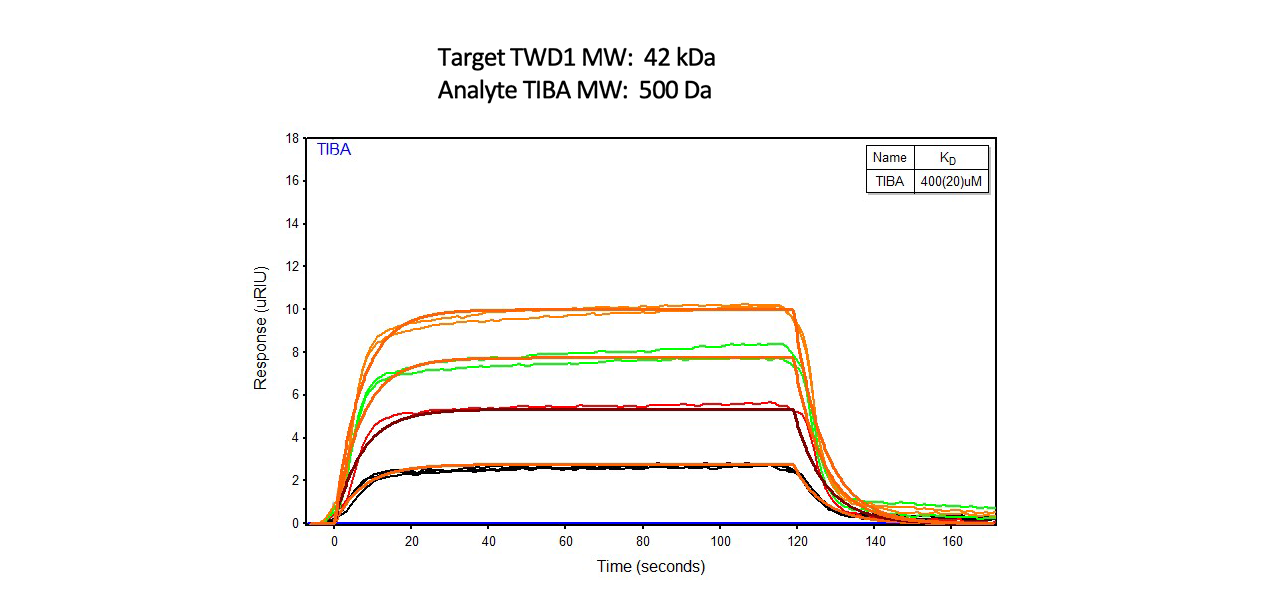
Example 8: Characterizing Inhibitors to Auxin Transport
Auxin is a plant growth regulator that has been widely studied. In a 2016 article in Plant Cell, researchers used a 2-channel Reichert SPR instrument to study binding interactions of several small molecule inhibitors with the protein Twisted Dwarf 1 (TWD1), which mediates the action of auxin transport inhibitors9. If a plant is grown on land, most of its development is dependenton the formation of auxin gradients called polar auxin transport or PAT. Synthetic auxin transport inhibitors, such as NPA (1-N-naphthylphthalamic acid), can inhibit the basal polar auxin flow required for plant development at low concentrations.
Using a 2-channel Reichert SPR, researchers immobilized TWD1 to the surface of a dextran chip using both amine and thiol coupling and flowed over small molecules at various concentrations. Using SPR, researchers determined that binding of small molecule inhibitors to the Twisted Dwarf protein was low affinity in vitro. This result was in agreement with NMR results9.
SPR binding results for one of the small molecule inhibitors is shown here:
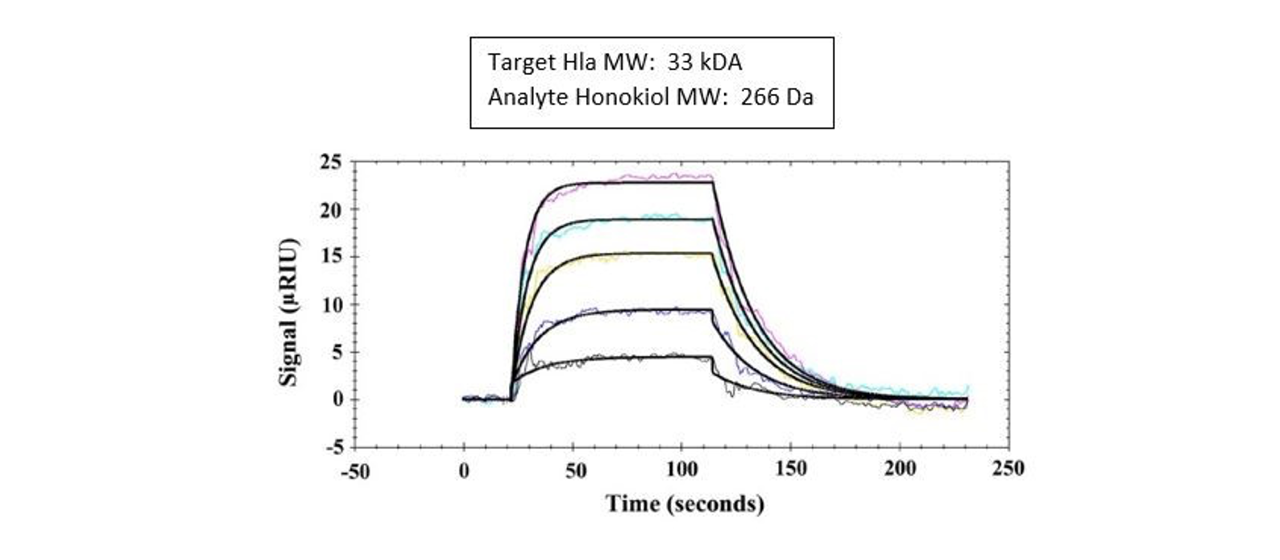
Example 9: Honokiol Inhibition of the pathogen Staphylococcus aureus (S. aureus)
In a 2019 article in Emerging Microbes and Infections, researchers used SPR to study the binding of Honokiol, a natural plant polyphenol which they determined to be a potential inhibitor of a common pathogen Staphylococcus aureus (S. aureus).10 Researchers used a Reichert4SPR to study this interaction. They immobilized α-Hemolysin (Hla) on a dextran chip and flowed over the Honokiol at various concentrations to determine the affinity.
Researchers in this study confirmed that α-Hemolysin (Hla), an extracellular toxin secreted by most pathogenic S. aureus strains, bound to Honokiol with a modest binding affinity. Additional research using other techniques showed that Honokiol, a low molecular weight natural product, can inhibit inflammasome activation without affecting oligomerization.10
The SPR binding curves are shown here:
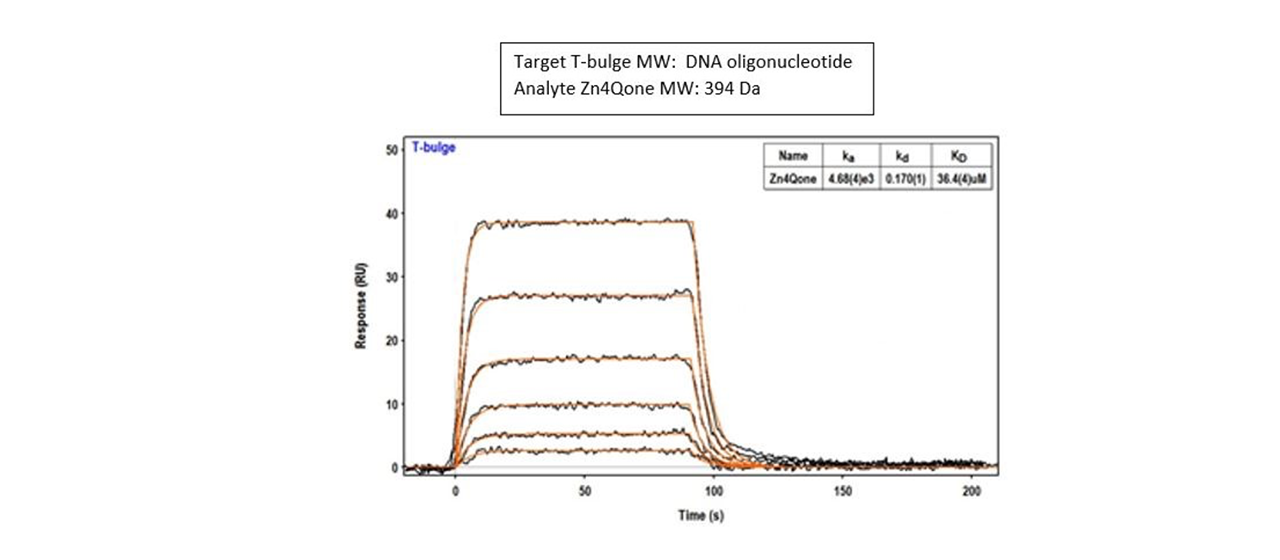
Example 10: Small molecule zinc complexes binding to DNA
In a 2015 article in Dalton Transactions, researchers designed small molecule zinc complexes and determined how they fit into bulges and quadraplexes on DNA. Non-canonical nucleobases that are part of DNA bulges are common targets when designing small molecules for binding both organic and inorganic compounds. And Zn(II) complexes containing planar aromatic pendents with two fused rings bind to DNA Hairpin (T-bulge) more tightly than complexes with non- planarpendents.11
Both a better understanding of these types of interactions and the development of alternative ways to recognize unusual nucleic acid structures could lead to improved design of small molecules for future therapeutic applications. Researchers in this study used Reichert’s 2-channel surface plasmon resonance (SPR) system to capture biotinylated DNA oligos on a planar streptavidin chip. They then flowed over the analytes at various concentrations to characterize the binding of Zn complexes to DNA T-bulge and T-loop structures.11
An example of a typical result is shown here:
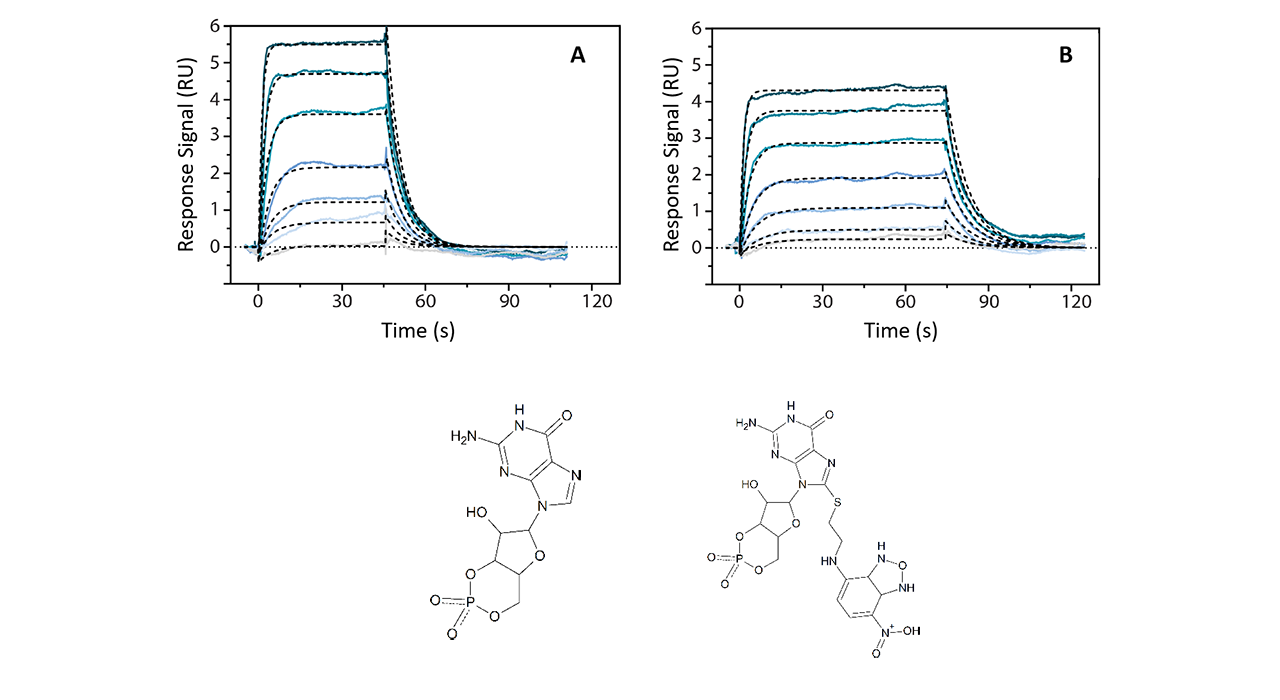
Example 11:
Most malaria deaths are caused by the protozoan parasite Plasmodium falciparum. Its life cycle is regulated by a cGMP-dependent protein kinase (PfPKG), inhibition of which is a promising antimalarial strategy. Allosteric kinase inhibitors, such as cGMP analogs, offer enhanced selectivity compared with competitive kinase inhibitors. However, the mechanisms underlying allosteric PfPKG inhibition are poorly understood. In this work, it was shown that 8-NBD-cGMP is an effective PfPKG antagonist12.
The challenging part of the experiments was the site-directed immobilization of PfPKG via its His6-tag while maintaining full functionality. Typically, small molecule interactions require covalent immobilization strategies as the baseline drift that normally occurs with Ni-NTA sensor chips is too big for the analysis of small signals during the analyte interaction. This problem was solved by using a XanTec high affinity polydentate Ni-NTA sensor chip (NiHC1000M) which binds His6-tagged ligands with extremely high stability so that barely any baseline drift occurs. Thus, the small molecule inhibitors cGMP and 8-NDB-cGMP were accessible for kinetic analysis. It was found that both had similar affinities (Kd 59 nM for cGMP and 51 nM for 8-NDB-cGMP). This example demonstrates that XanTec sensor chips are compatible with other SPR instruments (in this case a Biacore© T200), where they are an excellent means to boost the instrument performance.
Conclusion
SPR can play a pivotal role in development of therapeutics by the characterization of small molecule interactions with proteins. Reichert’s SPR systems have been implemented in a variety of research applications, and have the sensitivity and performance characteristics to meet challenging experimental needs such as low molecular weight interaction analysis – especially, when combined with sensor chips from XanTec bioanalytics.
Reichert offers two main SPR platforms – the 2SPR and 4SPR systems – which are affordable, flexible and have outstanding performance. Please contact us to learn more about how Reichert SPR can help you with the design and development of small molecule therapeutics.
For further information regarding XanTec’s range of SPR instruments, sensor chips and other consumables contact our team directly at sales@xantec.com.
References
- Shi H, Tice CM, Emert-Sedlak L, Chen L, Li WF, Carlsen M, Wrobel JE, Reitz AB, Smithgall TE. Tight-Binding Hydroxypyrazole HIV-1 Nef Inhibitors Suppress Viral Replication in Donor Mononuclear Cells and Reverse Nef-Mediated MHC-I Downregulation. ACS Infect Dis. 2020 Feb 14;6(2):302-312. doi: 10.1021/acsinfecdis.9b00382. Epub 2019 Dec 16. PMID: 31775511.
- Christian Brengel, Andreas Thomann, Alexander Schifrin, Giuseppe Allegretta, Ahmed A.M. Kamal, Jçrg Haupenthal, Isabell Schnorr, [ Sang Hyun Cho, Scott G. Franzblau, Martin Empting, Jens Eberhard, and Rolf W. Hartmann, „Biophysical Screening of a Focused Library for the Discovery of CYP121Inhibitors as Novel Antimycobacterials, “ ChemMedChem, 2017, 12, 1616 – 1626. DOI: 10.1002/cmdc.201700363
- Chuanbo Liu, Zuojia Liu and Jin Wang, “Uncovering the molecular and physiological processes of anticancer leads binding human serum albumin: A physical insight into drug efficacy, ” PLOS ONE April 20, 2017. doi.org/10.1371/journal.pone.0176208.
- BrookeL Brauer, Thomas M Moon, Sarah R Sheftic, Isha Nasa, Rebecca Page, Wolfgang Peti and Arminja N Kettenbach, “Leveraging new definitions of the LxVP SLiM to discover novel Calcineurin regulators and substrates, ” ACS Chem. Biol., 2019. DOI: 10.1021/ acschembio.9b00606
- Carolina Lixa, Michael W Clarkson, AnwarIqbal, Thomas M Moon, Fabio C L Almeida, Wolfgang Peti and Anderson S Pinheiro, “Retinoic acid binding leads to CRABP2 rigidification and dimerization, ” Biochemistry, 2019, Publication Date: September 30, 2019. doi.org/10.1021/acs.biochem.9b00672
- Benoît Bestgen, Irina Kufareva, Weiguang Seetoh, Chris Abell, Rolf W Hartmann, Ruben Abagyan, Marc Le Borgne, Odile Filhol, Claude Cochet, Thierry Lomberget and Matthias Engel, “2-Aminothiazole Derivatives as Selective Allosteric Modulators of the Protein Kinase CK2. 2. Structure-Based Optimization and Investigation of Effects Specific to the Allosteric Mode of Action, ” J. Med. Chem. 2019, 62, 1817−1836, DOI: 10.1021/acs.jmedchem.8b01765
- Reprinted (adapted) with permission from J.Med.Chem.2019, 62, 1817−1836, Publication Date: Feb 1, 2019, DOI: 10.1021/acs.jmedchem.8b01765. Copyright © 2019 American Chemical Society.
- Philip D Mosier, Meng-Jung Chiang, Zhengshi Lin, Yamei Gao, Bashayer Althufairi, Qibing Zhou, Faik Musayev, Martin K Safo, Hang Xie & Umesh R Desai, “Broad Spectrum Anti-Influenza Agents by Inhibiting Self-Association of Matrix Protein 1, ” Scientific Reports 6, Article number: 32340, 2016, doi:10.1038/srep3234
- Jinsheng Zhu, Aurelien Bailly, Marta Zwiewka, Valpuri Sovero, Martin Di Donato, Pei Ge, Jacqueline Oehri, Bibek Aryal, Pengchao Hao, Miriam Linnert, Noelia Inés Burgardt, Christian Lücke, Matthias Weiwad, Max Michel, Oliver H Weiergräber, Stephan Pollmann, Elisa Azzarello, Stefano Mancuso, Noel Ferro, Yoichiro Fukao, Céline Hoffmann, Roland, Wedlich-Söldner, Jirí Friml, Clément Thomas and Markus Geisler, “TWISTED DWARF1 mediates the action of auxin transport inhibitors on actin cytoskeleton dynamics, ” Plant Cell Advance Publication. Published on April 6, 2016, doi:10.1105/tpc.15.00726
- Na Guo, Zuojia Liu, Zhiqiang Yan, Zonghui Liu, Kun Hao, Chuanbo Liu & Jin Wang, “Subinhibitory concentrations of Honokiol reduce α-Hemolysin (Hla) secretion by Staphylococcusaureus and the Hla-induced inflammatory response by inactivating the NLRP3 inflammasome”, Emerging Microbes & Infections, 2019, 8:1, 707-716, DOI:10.1080/22221751.2019.1617643.
- Kevin E Siters, Stephanie A Sander, Jason R Devlin, and Janet R Morrow. „Bifunctional Zn (ii) complexes for recognition of non- canonical thymines in DNA bulges and G-quadruplexes.“ Dalton Transactions, 2015, Vol 44, pp. 3708–3716.
- Byun, J. A., Van, K., Huang, J., Henning, P., Franz, E., Akimoto, M., ... & Melacini, G. (2020). Mechanism of allosteric inhibition in the Plasmodium falciparum cGMP-dependent protein kinase. Journal of Biological Chemistry, jbc-RA120.