Surface Plasmon Resonance (SPR) can be used as an effective tool for screening many samples against a target or targets immobilized on a sensor chip.1-3 Often researchers do initial research showing that a certain molecule with a particular molecular backbone can act as an inhibitor to a given protein. They then follow up with analogs to see if efficacy can be improved by making structural changes to the original molecule including addition of other functional groups. In the example highlighted here, researchers at the University of Pittsburgh used SPR to help characterize inhibitor analogs and determine structure-activity relationships which ultimately could aid in the development of a new class of anti-retroviral drugs.1
Experimental
Background
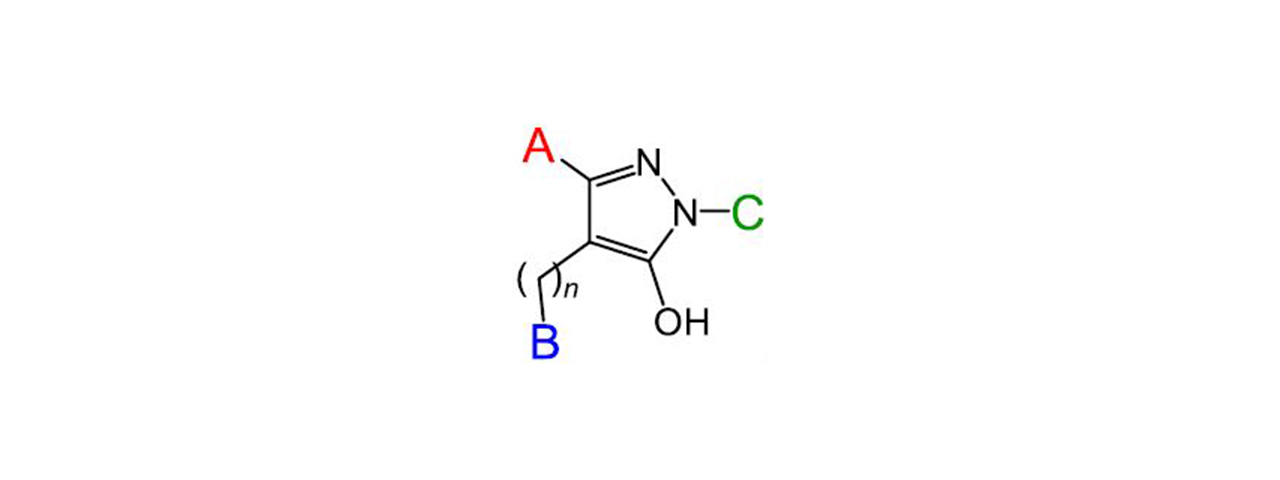
Earlier research identified a small molecule inhibitor of HIV- 1 Nef which was shown to block several of its activities related to enhancement of viral infectivity and replication.1 In the current research, 216 analogs of the original compound were synthesized and screened against recombinant Nef protein using SPR. The analogs all contained the same hydroxypyrazole scaffold from the original inhibitor (core molecule in the inset above) but with modifications at the A, B and C positions. From the SPR data obtained, compounds were compared based on affinity, extent of binding, and residence time (which is calculated from the reciprocal of the dissociation rate constant (1/koff)).1
Studying the SPR results, the top scoring inhibitors were found to share certain features, including a slow off rate. None of the top analogs returned to baseline over the course of a 3 minute dissociation, suggesting that these inhibitors might induce a change in the conformation of Nef that enables tight binding. However, no covalent bond was formed, since the analogs dissociated in weak base. The Pitt Researchers developed an “Activity Score” which combined multiple SPR parameters with anti- retroviral activity into a single numerical value for each analog providing an unbiased ranking of the compounds which helped them rank the analogs for further testing.1

Conditions
- Instrument: Reichert4SPR
- Sensor Chip: Carboxymethyl Dextran Temperature: 25°C
- Target: Recombinant, full-length HIV-1 Nef protein (NL4-3 allele)
- Analyte: Various
- Running Buffer: PBS + 1% DMSO
- Flow Rate: 50 μL/min
- Association Time: 1.5 minutes
- Dissociation Time: 3 minutes
- Regeneration: 5 mM NaOH for 30 seconds
Results
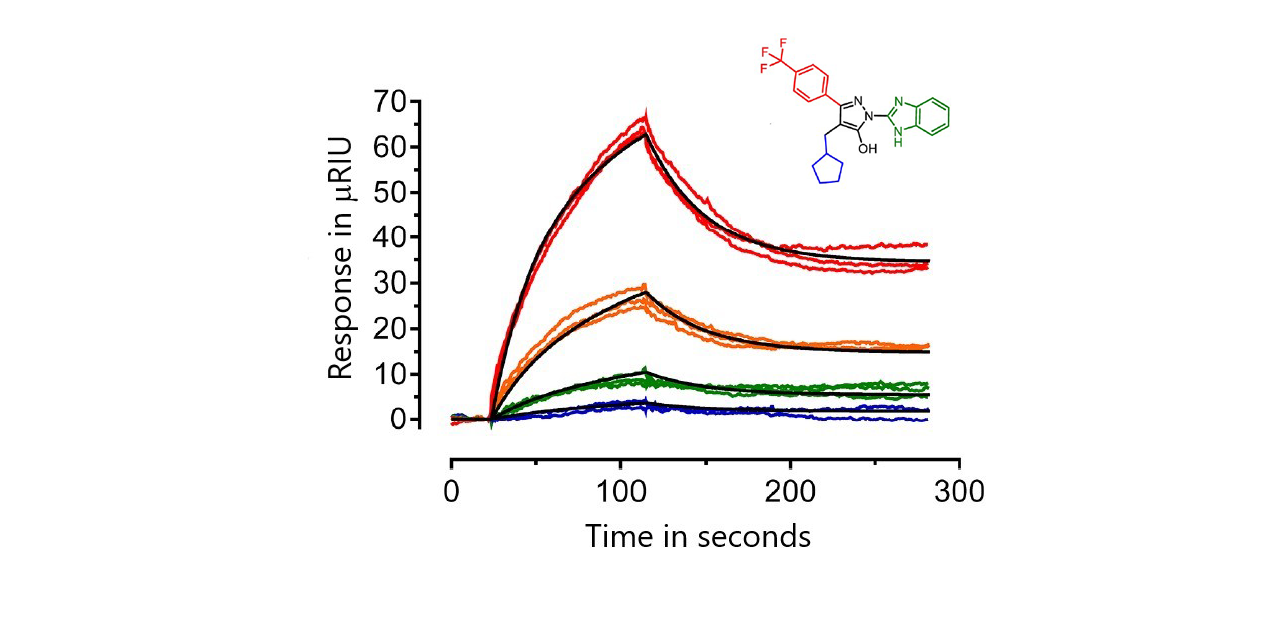
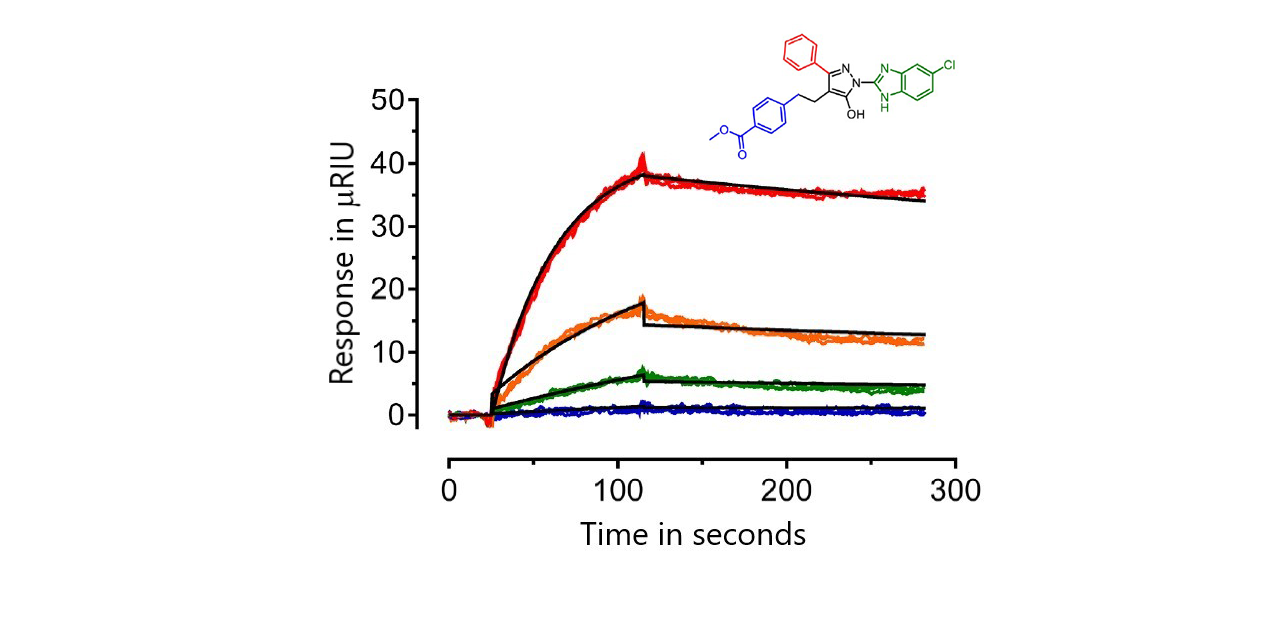
Forty-five of the analogs showed no binding. The remainder were ranked based on SPR kinetics, the size of the SPR response and their activity from a cell-based HIV-1 infectivity assay. Sensorgrams were fit to either a 1:1 model or a conformational change model. Affinities varied widely from low micromolar to picomolar. The top six analogs bound Nef with KD values ranging from about 0.65 pM (FC-7943) to 10 nM (FC-8698).1
Results for two of the top six compounds are shown here:
Summary
The screening example outlined here highlights how useful it is to do in vitro testing in real time using SPR. For this example, several hundred inhibitors were tested and ranked based on their kinetics and activity. By using SPR to help create an „Activity Score“, researchers were able to determine the top compounds to take into further testing. Their structure−activity approach led these researchers to identify new analogs with high affinity for Nef that also show strong antiretroviral activity and maintain the ability to reverse down regulation by Nef of MHC-I at the cell surface.1
References
- Shi H, Tice C M, Emert-Sedlak L, Chen L, Li W F, Carlsen M, Wrobel J E, Reitz A B, Smithgall T E. “Tight-Binding Hydroxypyrazole HIV-1 Nef Inhibitors Suppress Viral Replication in Donor Mononuclear Cells and Reverse Nef- Mediated MHC-I Downregulation”, ACS Infect Dis, 2020, 6, 302-312. doi:10.1021/acsinfecdis.9b00382.
- Christian Brengel, Andreas Thomann, Alexander Schifrin, Giuseppe Allegretta, Ahmed A. M. Kamal, Jçrg Haupenthal, Isabell Schnorr, Sang Hyun Cho, Scott G. Franzblau, Martin Empting, Jens Eberhard, and Rolf W. Hartmann, „Biophysical Screening of a Focused Library for the Discovery of CYP121Inhibitors as Novel Anti- mycobacterials,“ ChemMedChem, 2017, 12, 1616–1626. DOI:10.1002/cmdc.201700363
- Benoît Bestgen, Irina Kufareva, Weiguang Seetoh, Chris Abell, Rolf W. Hartmann, Ruben Abagyan, Marc Le Borgne, Odile Filhol, Claude Cochet, Thierry Lomberget and Matthias Engel, “2-Aminothiazole Derivatives as Selective Allosteric Modulators of the Protein Kinase CK2. 2. Structure-Based Optimization and Investigation of Effects Specific to the Allosteric Mode of Action,” J. Med. Chem. 2019, 62, 1817−1836, DOI: 10.1021/acs.jmedchem.8b01765
- Reprinted (adapted) with permission from Shi H, Tice C M, Emert-Sedlak L, Chen L, Li W F, Carlsen M, Wrobel J E, Reitz A B, Smithgall T E. “Tight-Binding Hydroxypyrazole HIV-1 Nef Inhibitors Suppress Viral Replication in Donor Mononuclear Cells and Reverse Nef-Mediated MHC-I Downregulation”, ACS Infect Dis, 2020, 6, 302-312. doi:10.1021/acsinfecdis.9b00382. Copyright © 2019 American Chemical Society.