Introduction
Surface plasmon resonance (SPR) has become an established technique for detection and quantification of biomolecular interactions. Sometimes, however, identification and structural elucidation is also needed. Proteolytic excision/extraction (Protex-MS), hydrogen-deuterium exchange (HDX-MS) of peptide backbone hydrogens, and Fast-Photochemical Oxidation (FPOP) are the main techniques used for mass spectrometry based elucidation of protein-ligand interactions, but none of these tools alone provide quantitative affinity data. The combination of SPR with Mass Spectrometry (MS) can be used to obtain both kinetics/affinity and identity/structural information about a biomolecular interaction.
A continuous online SPR-MS combination of a Reichert SPR instrument with an electrospray ionization mass spectrometer has been developed that enables the simultaneous affinity isolation, structure identification and affinity quantification of biopolymer ligands from a protein-ligand complex immobilized on a gold chip. A key part of the online SPR-MS epitope analyzer is an integrated, automated interface that provides sample concentration and in-situ desalting for the direct MS analysis of the ligand eluate [1]. ESI-MS systems from all major manufacturers can be coupled, using this newly developed software. The broad application potential of the online-SPRMS epitope analyzer has been shown by recent studies of an unusual mixed-disulfide antibody epitope of the rheumatoic target protein, HLA-B27; and the interaction site identification of chaperone complexes of lysosomal enzymes [2, 3]. Interaction epitopes as diverse as antigen-antibody and lectin-carbohydrate complexes [4], and affinity binding constants (KD) from milli- to nanomolar ranges can be detemined. Applications that can be studied using the online-SPR-MS epitope analyzer include affinity-based biomarker identification, identification of protein and peptide epitopes, precise antibody affinity determinations, and direct label free antigen quantification.
Experimental
To establish and validate the analytical feasibility and accuracy of the online SPRMS interface, direct analysis of horse heart myoglobin (HHM) by ESI-MS and SPR was carried out using a microfluidic interface. Myoglobin antibody was immobilized on a dextran-SPR affinity chip, as well as on a SepharoseTM micro-column. KD determination of myoglobin binding to myoglobin antibody was performed for the protein-antibody interaction using a Reichert SPR (2SPR) followed by ESI-MS analysis after sample processing through an interface. Subsequent online desalting of analyte prior to MS detection was performed after elution of affinity captured HHM from the affinity chip. Comparative data was recorded doing analysis of the protein in each separate instrument. Following the analysis of HHM and myoglobin antibody by both mass spectrometry and SPR analysis, the protein interaction and molecular identification were carried out by using the microfluidic interface shown in Figure 1.
Myoglobin is a well characterized protein, with molecular masses of 17558.132 Da or 16941.955 Da for holo- and apoprotein, respectively. Native HHM spectra were recorded under physiological pH in order to compare them to biosensor analysis conditions. Subsequent MS revealed denaturing sample processing conditions: HHM samples were eluted from the affinity column and desalted with 0.1 % TFA for ESI-MS analysis. The recorded spectra were those of the apo-myoglobin with charge distributions of [M+14H]14+ to [M+24H]24+ showing the unfolded, denatured protein (without heme). Kinetic studies of the affinity interaction from the biosensor with and without interface coupling were performed. After antibody immobilization, the KD values obtained were comparable whether they were obtained through direct analysis or in combination with on-line SPR-MS, proving the validity of the procedure.
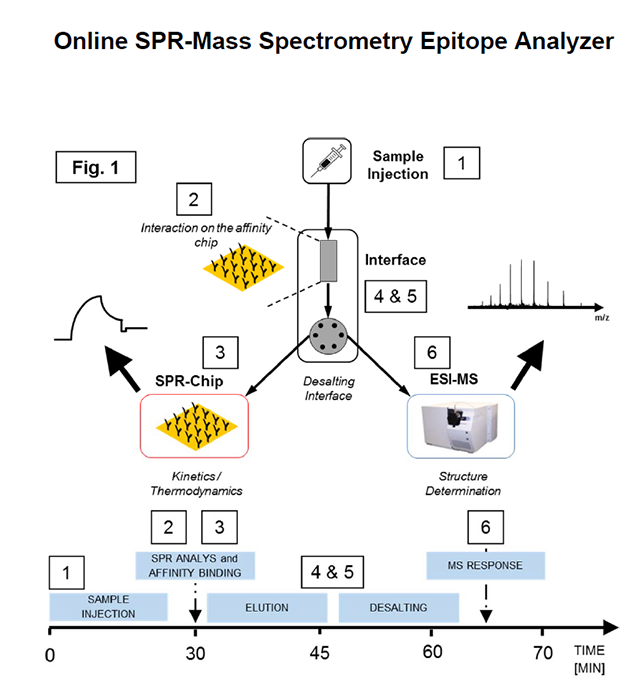
Schematic workflow of epitope & interaction analysis using the online SPR-MS Epitope Analyzer. After sample injection (1), the analyte is captured on the affinity chip (2) followed by the SPR-chip (3) for kinetic analysis of the affinity interaction. After sample processing through the desalting interface (4 and (5), structural analysis is performed by ESI-MS. Time scale is represented on the time axis below.
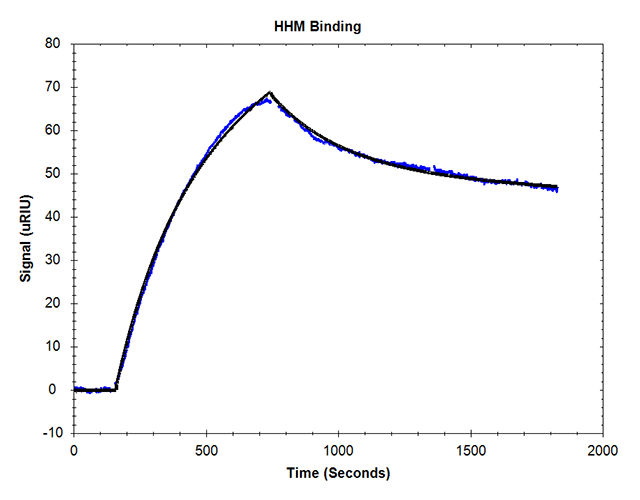
SPR kinetic evaluation of holo-myoglobin during online processing yielded the calculated KD of 450 nM.
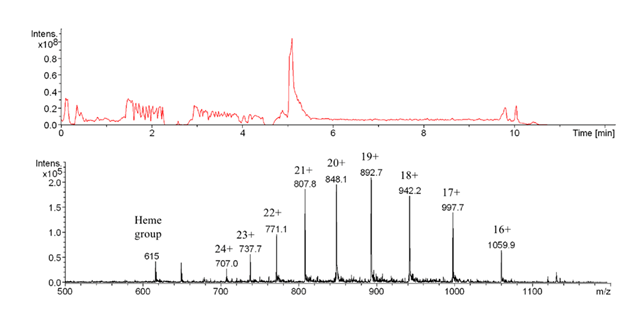
The total ion chromatogram shows a sharp elution signal around 5.3 min. The multiply charged protonated molecular ions identify the unfolded apoprotein recognized by the antibody (Bruker Esquire 3000+ ESI-MS).
Conclusions
The new online SRP-MS system is capable of detecting and identifying the affinity interaction of the myoglobin anti-myoglobin antibody pair in real time and the recorded spectra showed recognition of the apo-myoglobin. The reliability of the online interface was established by repeatable and comparable KD determinations and precise mass spectrometric identification of protein. Fast on-line sample processing allows fast throughput of different analytes for biomolecular interaction studies.
References
- Dragusanu, M., Petre, A., Slamnoiu, S., Vlad, C., Tu, T., Przybylski, M.(2010) J. Am. Soc. Mass Spectrom. 21: 1643-8.
- Iurascu, M.I., Marroquin Belaunzanar, O., Cozma, C., Petrausch, U., Renner C., Przybylski, M. (2016) J. Am. Soc. Mass Spectrom. 27: 1105-1112.
- Moise, A., Maeser, S., Rawer, S., Eggers, F., Murphy, M., Bornheim, J., Przybylski, M. (2016) J. Am. Soc. Mass Spectrom. 27: 1071-8.
- Moise A., Andre, S., Eggers, F., Krzeminski, M., Przybylski, M., Gabius, H.J. (2011) J. Am. Chem. Soc. 133, 14844-14847.