Serum IgG antibody binding to recombinant Ebola virus glycoprotein (GP) immobilized on carboxymethyl dextran surface
Introduction
This preliminary note involves the novel use of a Reichert SPR instrument to perform enrichment of phage antibodies using the fluidics system of the instrument. Phage antibody technology is a powerful method for obtaining specific antibodies directed at a defined target for a fraction of the time and cost of conventional monoclonal antibody technology. It was theorized that if phage encoding a binding partner for a target immobilized on a sensor slide could be separated from phage displaying non-binding proteins, then it would be possible to employ the precise fluidic controls of the instrument to perform enrichments under conditions that could prove beneficial in selecting affinity variants.
Experimental
The Reichert SPR instrument was used with a Reichert planar mixed SAM sensor chip and PBST as the running buffer. Approximately 1500 μRIU of BSA was amine-coupled to the left channel only of a two channel instrument. Phage displaying either an anti-BSA antibody (single-chain Fc, scFv) or an anti-peptide scFv (negative control) that can be easily distinguished by a colony PCR procedure were prepared and injected onto the sensor chip. Phage were applied at a flow rate of 10 μl/min. After 10 minutes, the flow rate was increased to 50 μl/min and aliquots of the outflow were collected. After approximately 20 minutes of dissociation, remaining sample was eluted by injection of 20 mM HCl and additional fractions collected. Phages present in the fractions were recovered by transduction of E. coli and samples of colonies in each fraction were subjected to colony PCR. The products were resolved by agarose gel electrophoresis.
Results
A summary of SPR and fractionation results are presented in Figures 1-3.
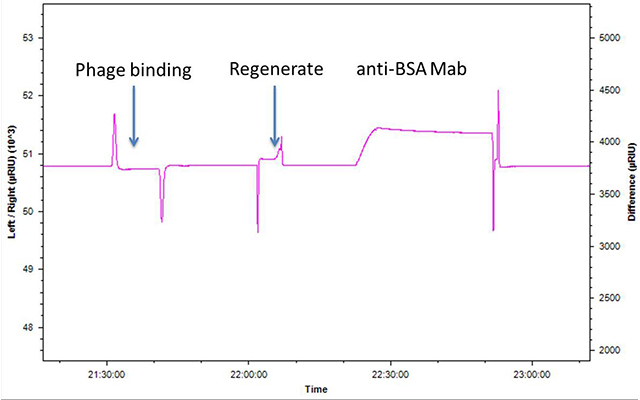
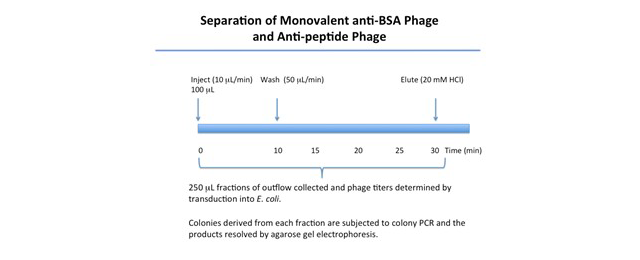
Conclusions
Preliminary results indicate that phage can be easily applied, fractionated and recovered from the SPR outflow after elution.
- Binding phage can be readily separated from non-binders using SPR. Results clearly show a 103 enrichment even though the system is not optimized.
- The SPR signal may be useful in characterization of panels of similar clones with varying affinities. It will be essential to use only monovalent phage. The yield of phage seems to be dependent on the target levels immobilized and affinity.
- Increasing the number of displaying phage in the input is desirable. The SPR signal is unlikely to be helpful when enriching for low abundance binders.
Potential opportunities/ Future directions
- Use SPR to characterize individual phage binding kinetics and to rank affinities
- Do binder selection using SPR affinity in phage. This would allow protein expression to be skipped.
- Use elution control to eliminate low affinity binders and improve reproducibility. Vary parameters such as flow rate, temperature, time, concentration and elution buffer composition depending on the relative affinities of the population.
- Immobilize different targets on different channels - this would allow for customization of specificity. Since there are multiple channels on the SPR, multiple enrichments could be run simultaneously.